Biologists decoded the structure of a long noncoding RNA, revealing its interaction with a cellular protein and its role in controlling the development of heart muscle cells.
New research details how biologists from MIT deciphered the structure of one type of long noncoding RNA and used that information to figure out how it interacts with a cellular protein to control the development of heart muscle cells.
Several years ago, biologists discovered a new type of genetic material known as long noncoding RNA. This RNA does not code for proteins and is copied from sections of the genome once believed to be “junk DNA.”
Since then, scientists have found evidence that long noncoding RNA, or lncRNA, plays roles in many cellular processes, including guiding cell fate during embryonic development. However, it has been unknown exactly how lncRNA exerts this influence.
Inspired by historical work showing that structure plays a role in the function of other classes of RNA such as transfer RNA, MIT biologists have now deciphered the structure of one type of lncRNA and used that information to figure out how it interacts with a cellular protein to control the development of heart muscle cells. This is one of the first studies to link the structure of lncRNAs to their function.
“Emerging data points to fundamental roles for many of these molecules in development and disease, so we believe that determining the structure of lncRNAs is critical for understanding how they function,” says Laurie Boyer, the Irwin and Helen Sizer Career Development Associate Professor of Biology and Biological Engineering at MIT and the senior author of the study, which appears in the journal Molecular Cell.
Learning more about how lncRNAs control cell differentiation could offer a new approach to developing drugs for patients whose hearts have been damaged by cardiovascular disease, aging, or cancer.
The paper’s lead author is MIT postdoc Zhihong Xue. Other MIT authors are undergraduate Boryana Doyle and Sarnoff Fellow Arune Gulati. Scott Hennelly, Irina Novikova, and Karissa Sanbonmatsu of Los Alamos National Laboratory are also the authors of the paper.
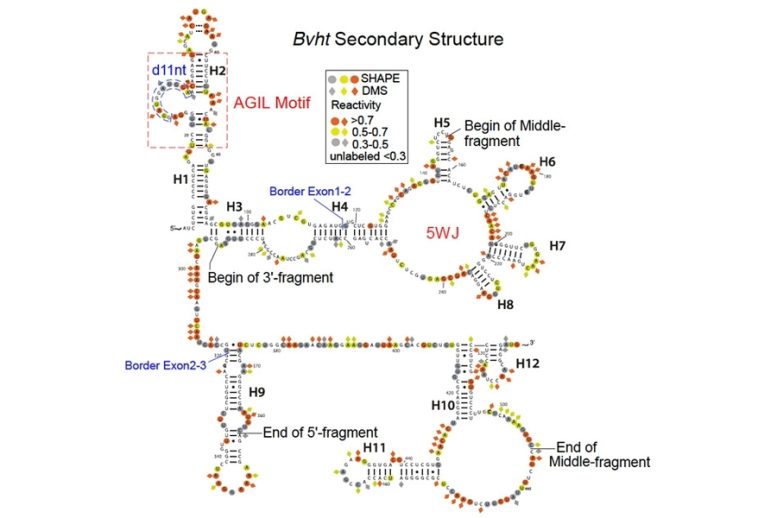
MIT biologists have deciphered the structure of a long noncoding RNA known as Braveheart. They found that the AGIL motif, at the top left, is critical to the molecule’s function.
Probing the heart
Boyer’s lab previously identified a mouse lncRNA known as Braveheart, which is found at higher levels in the heart compared to other tissues. In 2013, Boyer showed that this RNA molecule is necessary for the normal development of heart muscle cells.
In the new study, the researchers decided to investigate which regions of the 600-nucleotide RNA molecule are crucial to its function. “We knew Braveheart was critical for heart muscle cell development, but we didn’t know the detailed molecular mechanism of how this lncRNA functioned, so we hypothesized that determining its structure could reveal new clues,” Xue says.
To determine Braveheart’s structure, the researchers used a technique called chemical probing, in which they treated the RNA molecule with a chemical reagent that modifies exposed RNA nucleotides. By analyzing which nucleotides bind to this reagent, the researchers can identify single-stranded regions, double-stranded helices, loops, and other structures.
This analysis revealed that Braveheart has several distinct structural regions, or motifs. The researchers then tested which of these motifs were most important to the molecule’s function. To their surprise, they found that removing 11 nucleotides, composing a loop that represents just 2 percent of the entire molecule, halted normal heart cell development.
The researchers then searched for proteins that the Braveheart loop might interact with to control heart cell development. In a screen of about 10,000 proteins, they discovered that a transcription factor protein called cellular nucleic acid binding protein (CNBP) binds strongly to this region. Previous studies have shown that mutations in CNBP can lead to heart defects in mice and humans.
Further studies revealed that CNBP acts as a potential roadblock for cardiac development, and that Braveheart releases this repressor, allowing cells to become heart muscle.
“This is one of the first studies to relate lncRNA structure to function,” says John Rinn, a professor of stem cell and regenerative biology at Harvard University, who was not involved in the research.
“It is critical that we move toward understanding the specific functional domains and their structural elements if we are going to get lncRNAs up to speed with proteins, where we already know how certain parts play certain roles. In fact, you can predict what a protein does nowadays because of the wealth of structure-to-function relationships known for proteins,” Rinn says.
Building a fingerprint
Scientists have not yet identified a human counterpart to the mouse Braveheart lncRNA, in part because human and mouse lncRNA sequences are poorly conserved, even though protein-coding genes of the two species are usually very similar. However, now that the researchers know the structure of the mouse Braveheart lncRNA, they plan to analyze human lncRNA molecules to identify similar structures, which would suggest that they have similar functions.
“We’re taking this motif and we’re using it to build a fingerprint so we can potentially find motifs that resemble that lncRNA across species,” Boyer says. “We also hope to extend this work to identify the modes of action of a catalog of motifs so that we can better predict lncRNAs with important functions.”
The researchers also plan to apply what they have learned about lncRNA toward engineering new therapeutics. “We fully expect that unraveling lncRNA structure-to-function relationships will open up exciting new therapeutic modalities in the near future,” Boyer says.
Reference: “A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardiovascular Lineage” by Zhihong Xue, Scott Hennelly, Boryana Doyle, Arune A. Gulati, Irina V. Novikova, Karissa Y. Sanbonmatsu and Laurie A. Boyer, 8 September 2016, Molecular Cell.
DOI: 10.1016/j.molcel.2016.08.010







Non-coding DNA which translates to non-coding RNA was hitherto considered to be junk and now it assumes very great importance in the life formation. Non-coding DNA formed switches to regulate the tissue growth of specific organs.Now non coding DNA determines the fate of the tissue also . All tissues should undergo apoptosis or fail to die and this non coding DNA through incRNA (Non coding RNA) which is an interference RNA decides the fate of the cell. When cells don’t die it becomes cancerous. If the cell dies very soon it is a degenerative disease. Birth , Growth and Death are the regimen of each and every cell which should be ordained regularly in the life. This is an eye opener for the new vistas in the study of genetics. Thank You.