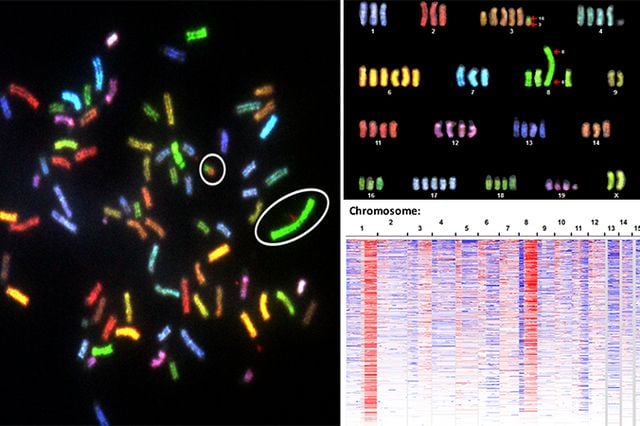
Genomically instable cancer cells are circled at left. Grouping like chromosomes together (right), helps scientists better detect the genetic aberrations that drive cancer growth.
A UCLA led study reveals that DNA patterns can unlock how glucose metabolism drives cancer.
FINDINGS
A UCLA-led study of DNA patterns in tumor cells suggests that the abundance of aberrant genetic signatures common in more aggressive cancers is not random but reflects selective forces in tumor evolution. The findings also demonstrate that these changes drive glucose metabolism in cells, which can lead to accelerated cancer growth.
BACKGROUND
Less aggressive cancers are known to have an intact genome — the complete set of genes in a cell — while the genome of more aggressive cancers tends to have a great deal of abnormalities. Modern research has focused largely on individual cancer genes, specifically oncogenes and tumor-suppressor genes, as the primary targets for DNA mutations and copy number alternations. Mutation or amplification in oncogenes can turn healthy cells into tumor cells. When tumor-suppressor genes are not working properly cells can grow out of control. However, scientists continue to observe many recurrent copy number alteration patterns in tumors that cannot be fully explained by these canonical cancer genes.
It has long been known that a fundamental difference between healthy cells and tumor-forming cells is reprogrammed cellular metabolism. An altered metabolism benefits a transformed cell in many ways, particularly in the ability to convert glucose into energy. This process, known as glycolysis, can fuel tumor growth the most-aggressive cancers.
METHOD
In the five-year study, Thomas Graeber, UCLA professor of molecular and medical pharmacology, and his colleagues used a cross-cancer analysis of copy number alterations data from human tumors, cancer cell lines and mouse models of cancer. The team analyzed samples from 15 cancer types and found that copy number alteration patterns of DNA amplification and deletion were predictive of both increased glycolytic activity and rapid growth of tumor cells. To understand how enzymes might play a role in this process, the scientists compared the copy number alteration patterns of human tumors and mouse models. They discovered 26 DNA regions with consistent genetic alterations, containing 11 enzymes in the glycolysis pathway as well as known cancer-driving genes.
The researchers next used an experimental immortalization system and genetic engineering to directly demonstrate that glycolytic enzymes can impact the copy number alteration signature. By sampling cells at different points in time, the researchers were able to show how these patterns evolve and thus shape the cancer genome.
IMPACT
With these findings, researchers can build improved cancer models and advance the development of therapeutics for all types of the disease.
“By focusing on such a large dataset and more than just known point mutations, we now better understand how subtle patterns of combined genetic alterations can affect glucose metabolism and are in sum key drivers of the most aggressive cancers,” Graeber said. “Scientists now have a whole new sandbox where we can test and develop the tools that will lead to new treatments for people fighting this disease.”
AUTHORS
The new study was led Graeber, the paper’s senior author and a member of the UCLA Jonsson Comprehensive Cancer Center’s Cancer Nanotechnology Program. The first authors are Nicholas Graham, assistant professor of chemical engineering at the USC Viterbi School of Engineering, and Aspram Minasyan, postdoctoral fellow in the UCLA Crump Institute of Molecular Imaging.
Additional co-authors include Anastasia Lomova, Ashley Cass, Nikolas Balanis, Michael Friedman, Shawna Chan, Sophie Zhao, Adrian Delgado, James Go, Lillie Beck, Rong Qiao, Johanna ten Hoeve, Nicolaos Palaskas, Hong Wu, Daniel Braas and Heather Christofk of UCLA. Graeber, Wu and Christofk are UCLA Jonsson Comprehensive Cancer Center members.
JOURNAL
The study was published online in Molecular Systems Biology.
FUNDING
The research was supported by the National Cancer Institute, National Institutes of Health, American Cancer Society, Melanoma Research Alliance, Norton Simon Research Foundation and Concern Foundation. Additional support was provided by UCLA’s Jonsson Cancer Center Foundation, UCLA CTSI, UC Cancer Research Coordinating Committee, UCLA Stein/Oppenheimer Endowment and private philanthropy.
Reference: “Recurrent patterns of DNA copy number alterations in tumors reflect metabolic selection pressures” by Nicholas A Graham, Aspram Minasyan, Anastasia Lomova, Ashley Cass, Nikolas G Balanis, Michael Friedman, Shawna Chan, Sophie Zhao, Adrian Delgado, James Go, Lillie Beck, Christian Hurtz, Carina Ng, Rong Qiao, Johanna ten Hoeve, Nicolaos Palaskas, Hong Wu, Markus Müschen, Asha S Multani, Elisa Port, Steven M Larson, Nikolaus Schultz, Daniel Braas, Heather R Christofk, Ingo K Mellinghoff and Thomas G Graeber, 15 February 2017, Molecular Systems Biology.
DOI: 10.15252/msb.20167159





Be the first to comment on "UCLA Study Shows DNA Patterns Can Unlock How Glucose Metabolism Drives Cancer"