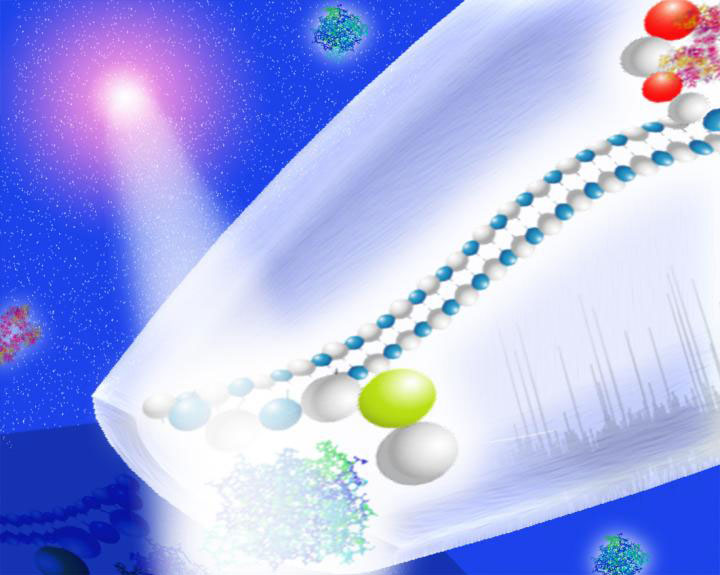
This is an illustration of the system in action. Credit: Joshua Edel/Imperial College London
The presence of, or changes in the concentration of, certain proteins in biological fluids can be indicators of disease. However, in the early stages of disease these ‘biomarkers’ can be difficult to detect, as they are relatively rare.
Detecting important biomarkers in lower concentrations will allow patients to be treated earlier for diseases such as some cancers and neurological disorders, which could increase the chance of survival rate.
However, current methods of detection are often not sufficiently sensitive and require costly and time-consuming sample pre-treatment.
Now, researchers from the Department of Chemistry at Imperial College London have come up with a system that is specific, flexible, and can detect single protein biomarkers directly in human serum (a pool of fluid separated from blood).
The system represents a significant innovation, as it is more sensitive to specific biomarkers and does not require clinical sample preparation. The method is published in Nature Communications.
Dr. Alex Ivanov, co-leader of this study from the Department of Chemistry at Imperial, said: “The detection of single molecules of biomarkers represents the ultimate in sensitivity for early diagnosis. We have now shown that this is possible to perform such measurements in real human samples, opening up the potential for meaningful early diagnosis.”
Binding to biomarkers
The method the team developed uses the ‘backbone’ of DNA, the structure it is built around. They grafted ‘aptamers’ – synthetic DNA molecules that bind to specific target biomarkers – to DNA backbones.
When added to human serum, the aptamers bind to biomarkers before being analyzed by passing through a nanopore detector. Nanopores are miniscule holes (often as small as a few billionths of a meter) that measure a change in electrical current as molecules pass through them.
Each biomarker has a unique current signature, so the presence and concentration of target biomarkers can be analyzed in this way.
The team demonstrated that their system can work by testing three aptamers on one DNA backbone. They found that the nanopores can detect the specific biomarkers that the aptamers were designed to pick up.
They say that the system can be constructed with more than five different aptamers, allowing detection of multiple biomarkers at once. In addition, the biomarkers were detected in human serum, meaning far less preparation time and cost were needed.
Based on the preliminary findings of this study, research efforts are now focused on several types of cancer and neurological disorders, which can benefit from the detection of biomarkers that are in low abundance in clinical samples.
The team have filed a patent for the technology, and are currently exploring routes towards commercialization so that it can be used to ultimately improve quality of life.
Dr. Jasmine Sze, who completed this study as part of her Ph.D. in the Department of Chemistry and has recently moved to the IMED Biotech at AstraZeneca said “Looking forward, with the rapid growth in nanotechnology and nanopore technology, this innovative platform could pave the way for the next wave of clinical applications.
“It has great potential for biomarker discovery, development of companion diagnostics as well as clinical endeavors, such as direct diagnosis, prognosis and sub-type classification with single-molecule sensitivity.”
Reference: “Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers” by Jasmine Y. Y. Sze, Aleksandar P. Ivanov, Anthony E. G. Cass and Joshua B. Edel, 16 November 2017, Nature Communications.
DOI: 10.1038/s41467-017-01584-3






Be the first to comment on "New Detection System Could Dramatically Improve Early Disease Diagnosis"