Therapeutic approach developed by Weizmann Institute scientists could spell new hope in the battle against COVID-19.
Even though vaccines may be steering the world toward a post-pandemic normal, a constantly mutating SARS-CoV-2 necessitates the development of effective drugs. In a new study published in Nature Microbiology, Weizmann Institute of Science researchers, together with collaborators from the Pasteur Institute, France, and the National Institutes of Health (NIH), USA, offer a novel therapeutic approach to combating the notorious virus. Rather than targeting the viral protein responsible for the virus entering the cell, the team of researchers addressed the protein on our cells’ membrane that enables this entry. Using an advanced artificial evolution method that they developed, the researchers generated a molecular “super cork” that physically jams this “entry port,” thus preventing the virus from attaching itself to the cell and entering it.

Prof. Gideon Schreiber. A new approach for treating COVID-19 developed by means of artificial evolution. Credit: Weizmann Institute of Science
Most potential therapies (and present vaccines) for COVID-19 target the so-called “spike protein” found on the virus’s outer envelope. This protein, however, is prone to mutations that erode the efficacy of these treatments. “Since the virus is constantly evolving we have, instead, focused on the non-evolving human receptor called ACE2 that acts as the entry site for the virus,” says Prof. Gideon Schreiber of Weizmann’s Biomolecular Sciences Department, who supervised the new study. This approach is not susceptible to new emerging virus variants, which is one of the main challenges in fighting the pandemic.
ACE2, attached to the membrane of lung epithelial cells and other tissues, is an enzyme important for regulating blood pressure. Therefore, as tempting as it may be to simply block this receptor to prevent the entry of SARS-CoV-2, any such strategy must not interfere with ACE2’s function. Schreiber, whose lab specializes in studying interactions between proteins, set out to develop a small protein molecule that could bind to ACE2 better than SARS-CoV-2 does but without affecting the receptor’s enzymatic activity.
The researchers, led by Dr. Jiří Zahradník, a postdoctoral fellow in Schreiber’s group, began by identifying SARS-CoV-2’s binding domain: the relatively short sequence of building blocks within the larger spike protein that physically binds to ACE2. Using the virus’s own receptor binding domain as a weapon against it, Zahradník performed several rounds of “evolution-in-a-test-tube,” developed in Schreiber’s lab, on a genetically engineered strain of baker’s yeast. Since yeast can be easily manipulated, Zahradník was able to rapidly scan millions of different mutations that accumulated in the course of this artificial evolution, a process that imitates natural evolution at a much faster pace. Ultimately, the goal was to find a small molecule that would be significantly “stickier” than the original viral version.
The rapid evolutionary process resulted in a small protein fragment with a binding capability 1,000 times stronger than that of the original viral binding domain
During this scanning process, Schreiber’s team supplied strong evidence in favor of the hypothesis that SARS-CoV-2 becomes more contagious when mutations improve its fit to ACE2. The researchers found that already after the first round of selection, the lab-made variants with tighter binding capabilities to ACE2 mimicked the mutations present in the binding domains of the most contagious SARS-CoV-2 strains that had occurred by means of natural evolution, such as the British variant (Alpha), the South African variant (Beta) and the Brazilian variant (Gamma). Surprisingly, the now widespread Indian (Delta) variant relies on a different trick to be more infectious – by partially evading detection by the immune system.
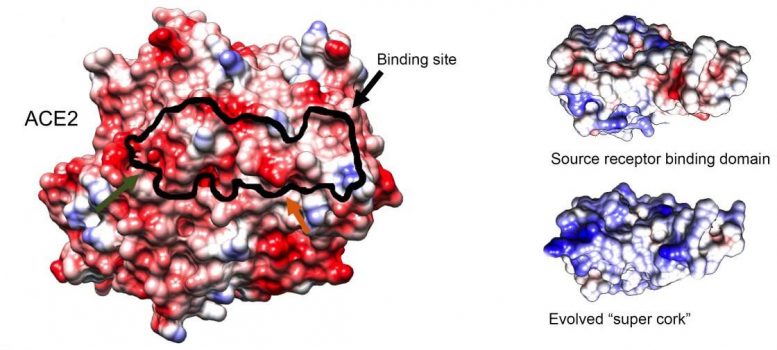
Structure of an ACE2 receptor (left), the original binding molecule (top right) and the newly engineered “super cork” (bottom right), imaged by cryogenic electron microscopy performed by Staff Scientists Dr. Nadav Elad of Weizmann’s Chemical Research Support Department and Dr. Orly Dym of the Life Sciences Core Facilities Department. The black outline indicates the binding site of the “super cork” on the ACE2 receptor. Credit: Weizmann Institute of Science
Eventually, Zahradník isolated a small protein fragment with a binding capability 1,000 times stronger than that of the original binding domain from which it evolved. This “super cork” not only fit ACE2 like a glove, it was also found by Maya Shemesh and Shir Marciano, PhD students in Schreiber’s lab, to conserve the enzymatic activity of ACE2 – just as the researchers had intended. Furthermore, owing to the strong binding, very low concentrations of the newly engineered molecule were needed to achieve the desired blocking effect.
While searching for a “super cork” that would block the ACE2 receptor, the researchers examined about 1,000,000,000 yeast mutants.
To develop a potential method for administering the molecule as a drug, Schreiber and his team collaborated with Prof. Yinon Rudich of Weizmann’s Earth and Planetary Sciences Department. Together with Dr. Ira Marton and Dr. Chunlin Li, they created an aerosol-based spray that would allow the developed molecule to be administrated by inhalation to patients.
So far, the researchers have tested, at the NIH, the developed formulation in hamsters infected with SARS-CoV-2. Preliminary results indicate that this treatment significantly reduces disease symptoms, suggesting that it could be a potential drug. More preclinical studies are planned to take place at the NIH in the near future.
Reference: “SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution” by Jiří Zahradník, Shir Marciano, Maya Shemesh, Eyal Zoler, Daniel Harari, Jeanne Chiaravalli, Björn Meyer, Yinon Rudich, Chunlin Li, Ira Marton, Orly Dym, Nadav Elad, Mark G. Lewis, Hanne Andersen, Matthew Gagne, Robert A. Seder, Daniel C. Douek and Gideon Schreiber, 16 August 2021, Nature Microbiology.
DOI: 10.1038/s41564-021-00954-4
Prof. Gideon Schreiber’s research is supported by the Ben B. and Joyce E. Eisenberg Foundation; the Rene and Tillie Molho Family Trust; Miel de Botton; and the Yotam Project.








Be the first to comment on "Putting a Super Cork on the Coronavirus: New Hope in the Battle Against COVID-19"