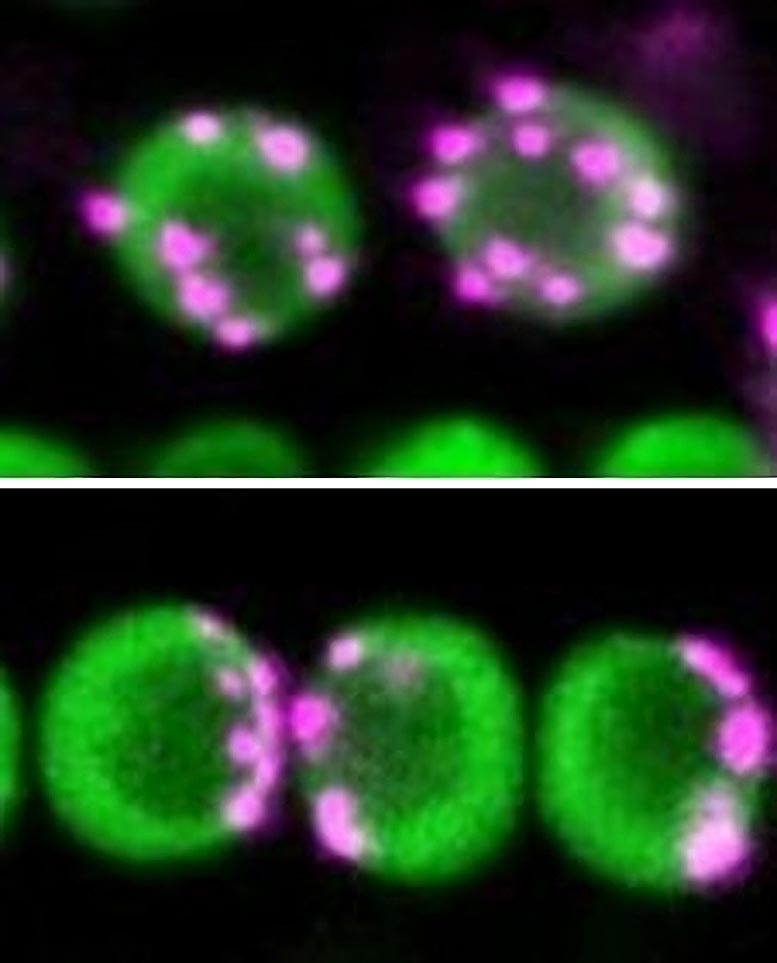
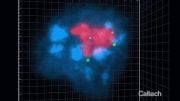
The upper panel shows non-Rabl configuration. Centromeres in magenta dispersed in nuclei in green. The lower panel shows Rabl configuration. Centromeres unevenly distributed in nuclei. Credit: Sachihiro Matsunaga, The University of Tokyo
Biologists uncover mechanism that shapes centromere distribution.
Since the 1800s, scientists have noted the configuration of centromeres, a special chromosomal region that is vital for cell division, in the cell nucleus. However, up until now, the determining mechanisms and the biological significance of centromere distribution were poorly understood. Recently, researchers proposed a two-step regulatory mechanism that shapes centromere distribution. Their findings also indicate that centromere configuration in the nucleus plays a role in maintaining genome integrity.
The results were published today (August 1, 2022) in the journal Nature Plants. The study was led by researchers from the University of Tokyo and their collaborators.
Special chromosomal domains known as centromeres are pulled to the opposite ends of the cell during the process of cell division. After cell division is complete and the cell nucleus is constructed, centromeres are spatially distributed in the nucleus. If the distribution of centromeres pulled to the two poles remains unchanged, the cell nucleus will have centromeres grouped at just one side of the nucleus. This uneven distribution of centromeres is called Rabl configuration, after Carl Rabl, the 19th-century cytologist. Some species’ nuclei show a dispersed distribution of centromeres instead. This is known as non-Rabl configuration.
“The biological function and molecular mechanism of the Rabl or non-Rabl configuration has been a mystery across the centuries,” said corresponding author Sachihiro Matsunaga, professor at the University of Tokyo’s Graduate School of Frontier Sciences. “We successfully revealed the molecular mechanism to construct the non-Rabl configuration.”
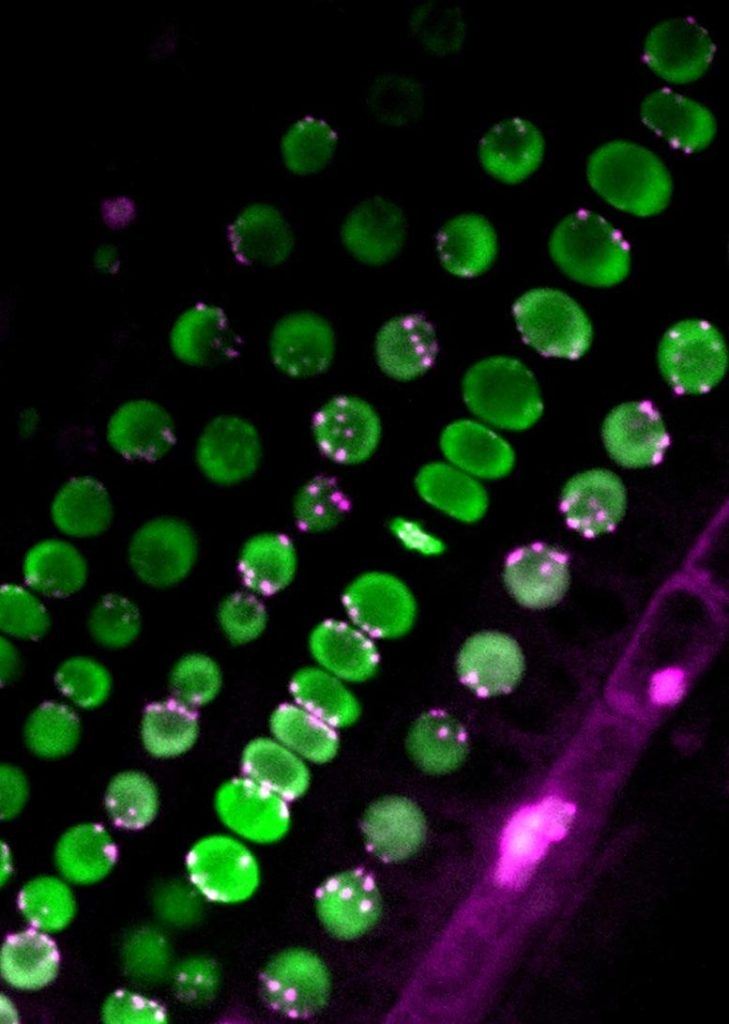
The uneven distribution of centromeres (magenta) in nuclei (green). Credit: Sachihiro Matsunaga, The University of Tokyo
The scientists studied the plant Arabidopsis thaliana, known also as thale cress and a specimen that is known to have non-Rabl configuration, and its mutant form that had a Rabl configuration. Through their work, they found that protein complexes known as condensin II (CII) and protein complexes known as the linker of nucleoskeleton and cotoskeleton (LINC) complex work together to determine centromere distribution during cell division.
“The centromere distribution for non-Rabl configuration is regulated independently by the CII– LINC complex and a nuclear lamina protein known as CROWDED NUCLEI (CRWN),” Matsunaga said.
The first step of the two-step regulatory mechanism of centromere distribution that the researchers uncovered was that the CII-LINC complex mediates the scattering of centromeres from late anaphase to telophase — two phases at the end of cell division. The second step of the process is that the CRWNs stabilize the scattered centromeres on nuclear lamina within the nucleus.
Next, to explore the biological significance, the researchers analyzed the gene expression in A. thaliana and in its Rabl-structure mutant. Because a change in the spatial arrangement of centromeres also changes the spatial arrangement of genes, the researchers expected to find differences in gene expression, but this hypothesis proved to be incorrect. However, when DNA damage stress was applied, the mutant grew organs at a slower rate than the normal plant.
“This suggests that precise control of centromere spatial arrangement is required for organ growth in response to DNA damage stress, and there is no difference in tolerance to DNA damage stress between organisms with the non-Rabl and Rabl,” Matsunaga said. “This suggests that the appropriate spatial arrangement of DNA in the nucleus regardless of Rabl configuration is important for stress response.”
According to Matsunaga, the next steps are to identify the power source that changes the spatial arrangement of specific DNA regions and the mechanism that recognizes specific DNA.
“Such findings will lead to the development of technology for artificially arranging DNA in the cell nucleus in an appropriate spatial arrangement,” he said. “It is expected that this technology will make it possible to create stress-resistant organisms, as well as to impart new properties and functions by altering the spatial arrangement of DNA rather than editing its nucleotide sequence.”
Reference: “Two-step regulation of centromere distribution by condensin II and the nuclear envelope proteins” by Takuya Sakamoto, Yuki Sakamoto, Stefan Grob, Daniel Slane, Tomoe Yamashita, Nanami Ito, Yuka Oko, Tomoya Sugiyama, Takumi Higaki, Seiichiro Hasezawa, Maho Tanaka, Akihiro Matsui, Motoaki Seki, Takamasa Suzuki, Ueli Grossniklaus and Sachihiro Matsunaga, 1 August 2022, Nature Plants.
DOI: 10.1038/s41477-022-01200-3



The changes occuring are not random but have a specific goal.
There is nothing to relate that any change occuring in cromosom orientation is to restòre the effecting fore as to gain modification is a natural process.