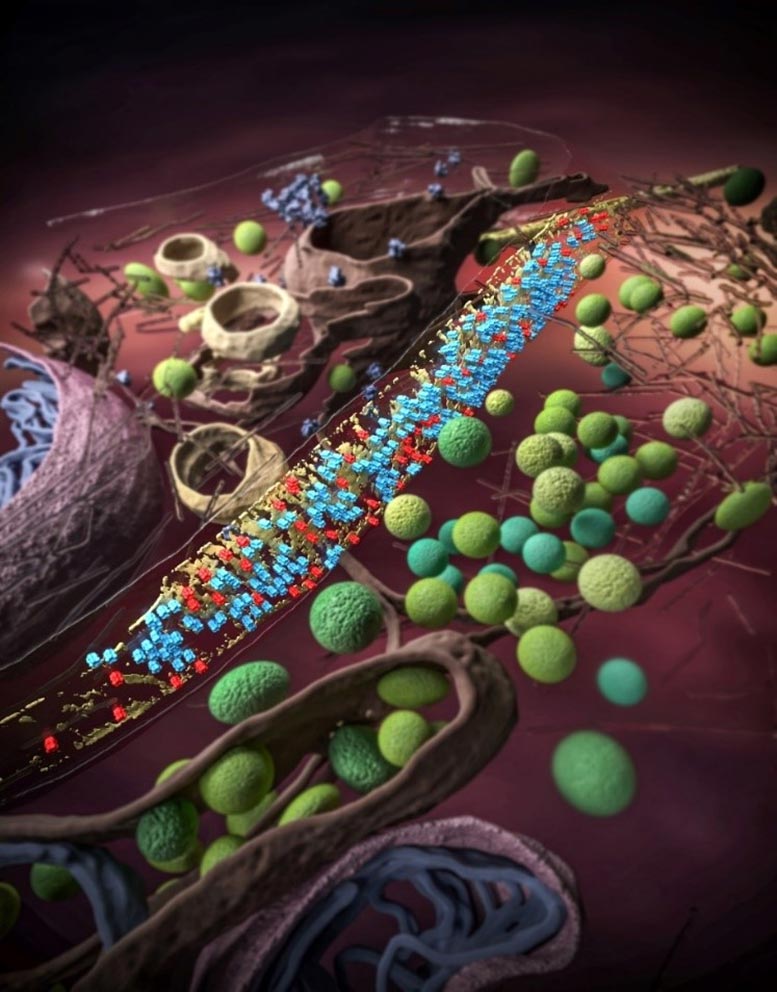
Synapses are specialized devices where learning and memory occur. The efficient transmission of synaptic signals relies on the delicate structure and complex molecular composition of the synapses. However, the small size (several hundred nanometers in diameter) and heterogeneous nature of the synapses pose significant challenges in direct observation of the molecules inside synapses.
Based on the proposed processing technique for in situ cryo-electron tomography, researchers from University of Science and Technology of China (USTC) and Shenzhen Institutes of Advanced Technology (SIAT) of the Chinese Academy of Sciences (CAS) became the first scientists to observe individual GABAA receptors and their organization on the synaptic membrane, endowing the brain’s ability for information processing.
“The advance of this study comes from the in situ cryo-electron microscopy, a method that preserves the cells in native states and has an order of magnitude of higher resolution compare to the super-resolution optical microscopy,” said TAO Changlu, postdoctoral fellow from USTC and the study’s co-first author, now associate investigator at SIAT.
This image processing technique is able to automatically locate the membrane proteins in their cellular context. “To ensure that we detect every receptor on the postsynaptic membrane, we oversampled the synaptic membrane and classified all the sampled 3D images without any template,” said LIU Yuntao, a graduate student from USTC and the study’s co-first author, now a postdoctoral fellow at UCLA. “We even used the negative control that sampling the presynaptic membrane to validate our observation.”
Once the receptors were detected, researchers suddenly realized that the receptors are not randomly distributed on the membrane: they tend to keep the same 11-nm “social distancing” from each other. Intriguingly, the receptors can rotate freely, even though constrained by distance.
“The social distancing among receptors could arise from their interactions with scaffolding molecule — gephyrins,” said BI Guoqiang, professor of neuroscience at USTC and senior author of the paper.
The scaffolding molecules form a 5-nm thick density sheet to support and regulate GABAA receptors on the membrane. Together, they form an absorbing semi-ordered structure called “mesophasic assembly.”
A mesophasic state is in between the liquid and solid, which might be induced by the multivalent interaction between receptors and their scaffolding molecules and attract the readily releasable vesicles containing neurotransmitters. The inhibitory synapses could store information by arranging the GABAA receptors in such a low-entropy Goldilocks state.
This semi-ordered structure differs from the previously proposed hexagonal lattice organization of GABAA receptors and gephyrins. Notably, each synapse tends to contain one mesophasic assembly, rather than multiple nano-domains as observed in excitatory synapses with super-resolution optical microscopy.
“This work represents the first nanometer-resolution observation at the inhibitory synaptic receptors and a critical step towards resolving the atomic details of the brain,” said ZHOU Hong, director of the Electron Imaging Center for NanoMachines at the California NanoSystems Institute at UCLA, also senior author of the paper.
This work is published in the journal Nature Neuroscience.
Reference: “Mesophasic organization of GABAA receptors in hippocampal inhibitory synapses” by Yun-Tao Liu, Chang-Lu Tao, Xiaokang Zhang, Wenjun Xia, Dong-Qing Shi, Lei Qi, Cheng Xu, Rong Sun, Xiao-Wei Li, Pak-Ming Lau, Z. Hong Zhou and Guo-Qiang Bi, 2 November 2020, Nature Neuroscience.
DOI: 10.1038/s41593-020-00729-w
Funding: National Natural Science Foundation of China, Chinese Academy of Sciences, National Key R&D Program of China. Tao was supported by the China Postdoctoral Science Foundation and the Natural Science Foundation of Anhui Province, National Institutes of Health.
Never miss a breakthrough: Join the SciTechDaily newsletter.
1 Comment
Did time, chance, and mutations produced the synapses as illustrated in the graphic above? Impossible! The complexity in this and billions of other biological mechanisms could only be created by a Mind beyond anything we can comprehend. We see His handiwork everywhere, and yet those in the “know” continue to credit mindless evolution for such advanced structures. Does one find even a simple arrow in the woods and not know that someone produced it? How is it that LIVING structures of such intricate detail can be assigned to anything other than a Creator?