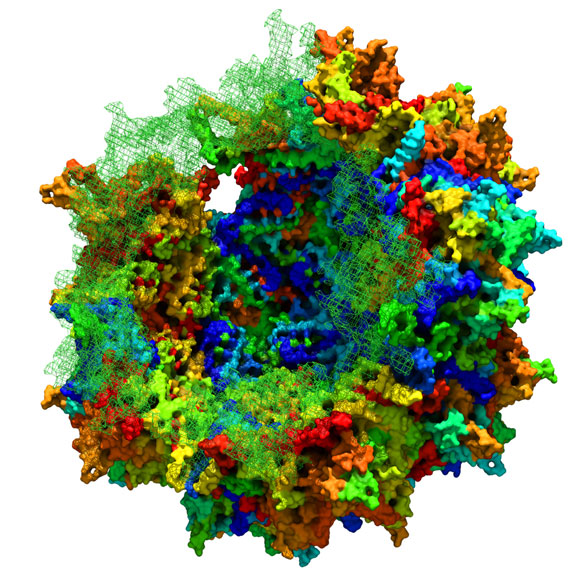
A cartoon depicting Adeno-associated viral capsid in formation by ancestral sequence reconstruction. Credit: Eric Zinn.
A team of researchers from Harvard Medical School has recreated the evolutionary lineage of adeno-associated viruses, allowing them to better understand the intricate structure of viruses and how different properties arose throughout evolution.
Harvard Medical School researchers have recreated the evolutionary lineage of adeno-associated viruses (AAVs) to reconstruct an ancient viral particle that is highly effective at delivering gene therapies targeting the liver, muscle, and retina.
This approach, published July 30 in Cell Reports, could be used to design a new class of genetic drugs that are safer and more potent than those currently available.
“Our novel methodology allows us to understand better the intricate structure of viruses and how different properties arose throughout evolution,” said senior study author Luk Vandenberghe, HMS assistant professor of ophthalmology at Mass. Eye and Ear.
“We believe our findings will teach us how complex biological structures, such as AAVs, are built. From this knowledge, we hope to design next-generation viruses for use as vectors in gene therapy,” he said.
Viruses need to efficiently transfer their genetic material into host cells in order to replicate and survive. Researchers have taken advantage of this natural property to develop viral vectors, or carriers, capable of shuttling therapeutic genes to the appropriate cells or tissues.
Early-stage clinical trials have demonstrated the safety and effectiveness of this approach for treating inherited blindness and hemophilia. But so far, AAVs used for gene therapy have been chosen from naturally circulating viral strains that patients may already have been exposed to, meaning they would have natural immunity. Because natural immunization blocks the transfer of the therapeutic gene, these individuals are often ineligible for gene therapy.
Generating AAV vectors that reliably evade the host immune system is pivotal for the long-term success of this promising therapeutic approach. However, efforts to engineer improved AAVs have been stymied by the intricate structure of these viruses. Like pieces of a jigsaw puzzle, every protein in the shell of the virus must fit together perfectly for the virus to function normally. Altering proteins in one part of the virus to achieve a certain benefit, such as more efficient gene transfer or reduced recognition by host immune cells, could end up destroying the structural integrity of the entire shell.
To overcome this challenge, Vandenberghe and his team at the Grousbeck Gene Therapy Center at MEE turned to evolutionary history for guidance. Under selective pressure, AAV ancestors have undergone a series of changes that altered viral function while preserving structural integrity and other core viral functions. “Analysis of these evolutionary intermediates provides insights into how to uncouple important vector properties to build safer and improved gene therapy vectors,” said first author Eric Zinn, research assistant at the Schepens Eye Research Institute.
Using computational techniques, the researchers analyzed amino acid sequences of contemporary AAVs and used this information to predict the ancestral amino acid sequence for the AAV protein shell. This analysis allowed them to reconstruct in the laboratory nine synthetic predecessors for AAVs that are currently undergoing clinical testing. The most ancient of those, named Anc80, was extensively tested for applications in gene therapy. When injected into mice, Anc80 successfully targeted the liver, muscle, and retina with levels of gene transfer equivalent or superior to those of contemporary AAVs used in clinical trials. Moreover, the ancestral virus did not produce any toxicity and was less susceptible to the immunity induced by AAVs currently circulating in nature. “Our work is the first demonstration of a computationally designed and fully synthetic viral vector for gene therapy,” Vandenberghe said.
According to Vandenberghe, generating novel infectious agents may raise some concerns, but several safety measures have been taken. For example, AAVs are not known to cause any diseases, and their replication machinery is crippled prior to their use in gene therapy.
In future studies, the researchers will characterize the interplay between the virus and host through evolution and continue to seek improved vectors for clinical applications. They will also examine the potential of Anc80 for treating liver diseases and retinal forms of blindness. “The vectors developed and characterized in this study demonstrate unique and potent biology that justify their consideration for gene therapy applications,” Vandenberghe said.
This work was supported by the Grousbeck Family Foundation, the Candyce Henwood Fund, the NIH Common Fund, Research to Prevent Blindness, and the Foundation Fighting Blindness.
Reference: “In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector” by Eric Zinn, Simon Pacouret, Vadim Khaychuk, Heikki T. Turunen, Livia S. Carvalho, Eva Andres-Mateos, Samiksha Shah, Rajani Shelke, Anna C. Maurer, Eva Plovie, Ru Xiao and Luk H. Vandenberghe, 30 July 2015, Cell Reports.
DOI: 10.1016/j.celrep.2015.07.019







Be the first to comment on "Biologists Recreate the Evolutionary Lineage of Adeno-Associated Viruses"