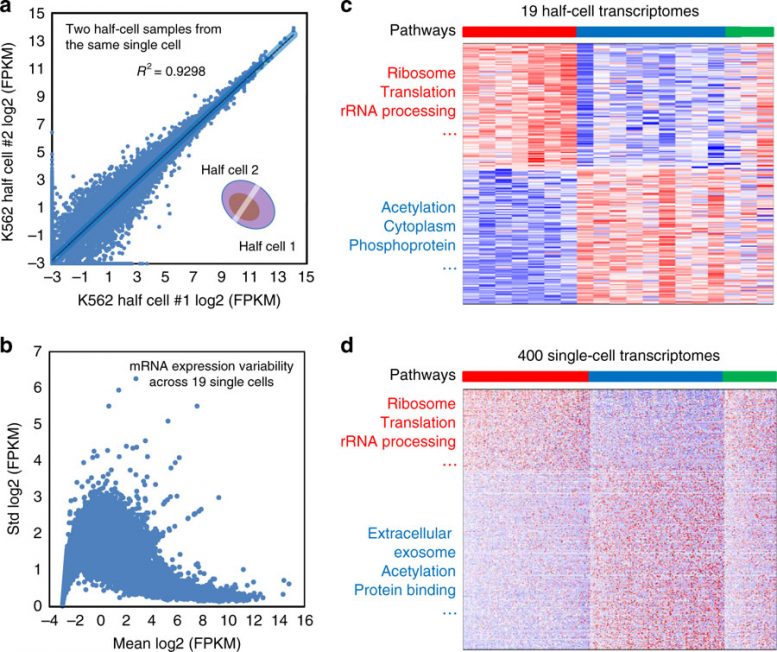
Global profiling of mRNAs from half cells. a A single K562 cell was lysed, and two halves of the lysate were separately subjected to RNAseq. Scatter plot of normalized and log2-transformed mRNA expression levels (in FPKM units) is shown. Each circle represents one annotated mRNA. The correlation coefficient R2 is 0.930. b The standard deviations of mRNA expression across 19 successfully profiled half cells were plotted against the mean expression, using log2-transformed mRNA expression data. Each circle represents one annotated mRNA. c Consensus clusters were identified in the 19 K562 half-cell mRNA expression data. A heatmap is shown for differentially expressed genes between the red and blue clusters, with enriched pathways annotated by the DAVID pathway analysis. Each row represents one annotated gene whereas each column represents a single cell. d K562 cells were profiled using a massively parallel single-cell 3′-end RNAseq technology. A heatmap is shown for differentially expressed genes between the red and blue consensus clusters, with enriched pathways annotated by the DAVID pathway analysis. Each row represents one annotated gene whereas each column represents a single cell. Within the heatmaps, blue color indicates low expression whereas red indicates higher expression
MicroRNAs, the tiny bits of genetic material that regulate gene expression, play a significant – but poorly understood – role in controlling the differences between individual cells. Yale researchers have developed a technology that sheds light on the workings responsible for these differences – a breakthrough that could lead to new insights about cancer development.
Researchers at Yale were able to profile for the first time both microRNA and messenger RNA (mRNA) in the same individual cell. By doing so, they discovered new mechanisms that lead to the heterogeneity – that is, the many variations – of gene expression. The results are published today in Nature Communications.
The research is a collaboration between the laboratories of Rong Fan, associate professor of biomedical engineering, and Jun Lu, an associate professor of genetics at the Yale Stem Cell Center.
“Our work for the first time allowed for the direct measurement of all microRNAs and all mRNAs in single cells, which can provide direct evidence of how microRNAs control their target mRNAs,” Fan said.
To co-sequence microRNA and mRNA from the same cell, the researchers needed to develop a new approach, since conventional methods, such as magnetic capture and gel electrophoresis, don’t work reliably at the single-cell scale. The researchers used a “half-cell genomics” approach. To do so, they physically split a single-cell lysate – that is, the liquid containing all the materials from a single cell – into two equal fractions. This is trickier than it sounds, since ensuring that all the protein-bound nucleic acid materials are fully dissociated is a complicated process.
For the study, the researchers showed that the half-cell approach could lead to robust splitting of microRNA and mRNA from single-cell lysates and that they could perform a co-measurement of both from the same cell. In doing so, they found evidence that variability in microRNA expression alone is responsible for non-genetic cell-to-cell heterogeneity.
The innovation could lead to unraveling numerous mysteries of the genome.
“This work brings new opportunities to investigate how the changes in small RNA expression in single cells contribute to non-genetic cell-to-cell variability, and how miRNA expression heterogeneity contributes to the control of single-cell level mRNA expression heterogeneity,” said Nayi Wang, a former Ph.D. student in Fan’s laboratory and lead author of the study.
The breakthrough could lead to a better understanding of cancer development, intratumor heterogeneity, and therapeutic resistance. The researchers said the next step may be to seek microRNA targets for cancer treatments.
Reference: “Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation” by Nayi Wang, Ji Zheng, Zhuo Chen, Yang Liu, Burak Dura, Minsuk Kwak, Juliana Xavier-Ferrucio, Yi-Chien Lu, Miaomiao Zhang, Christine Roden, Jijun Cheng, Diane S. Krause, Ye Ding, Rong Fan and Jun Lu, 9 January 2019, Nature Communications volume 10, Article number: 95 (2019).
DOI: 10.1038/s41467-018-07981-6





Be the first to comment on "Breakthrough Could Lead to New Insights About Cancer Development"