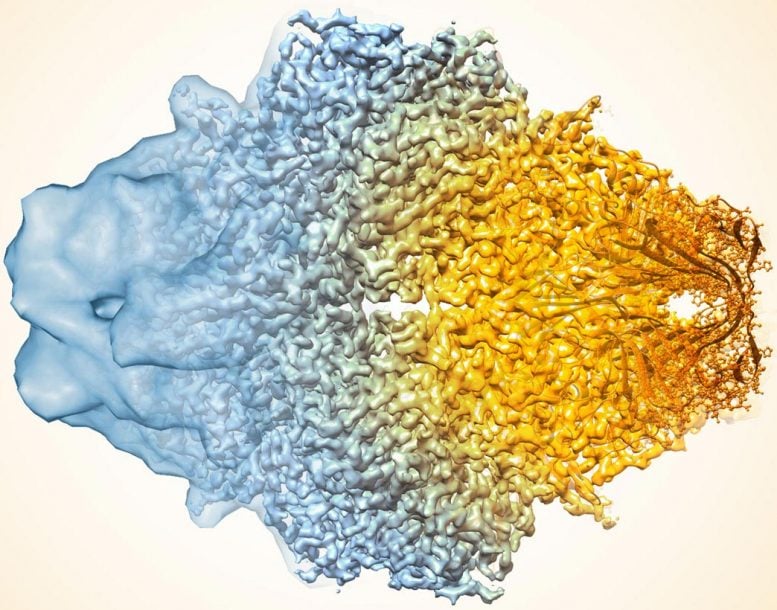
A composite image of the enzyme lactase showing how cryo-EM’s resolution has improved dramatically in recent years. Older images to the left, more recent to the right. Credit: Veronica Falconieri/National Cancer Institute
Scientists develop a technique that improves the resolution of cryo-electron microscopy.
We’ve all seen that moment in a cop TV show where a detective is reviewing grainy, low-resolution security footage, spots a person of interest on the tape, and nonchalantly asks a CSI technician to “enhance that.” A few keyboard clicks later, and voila — they’ve got a perfect, clear picture of the suspect’s face. This, of course, does not work in the real world, as many film critics and people on the internet like to point out.
However, real-life scientists have recently developed a true “enhance” tool: one that improves the resolution and accuracy of powerful microscopes that are used to reveal insights into biology and medicine.
In a study published in Nature Methods, a multi-institutional team led by Tom Terwilliger from the New Mexico Consortium and including researchers from Lawrence Berkeley National Laboratory (Berkeley Lab) demonstrates how a new computer algorithm improves the quality of the 3D molecular structure maps generated with cryo-electron microscopy (cryo-EM).
For decades, these cryo-EM maps — generated by taking many microscopy images and applying image-processing software — have been a crucial tool for researchers seeking to learn how the molecules within animals, plants, microbes, and viruses function. And in recent years, cryo-EM technology has advanced to the point that it can produce structures with atomic-level resolution for many types of molecules. Yet in some situations, even the most sophisticated cryo-EM methods still generate maps with lower resolution and greater uncertainty than required to tease out the details of complex chemical reactions.
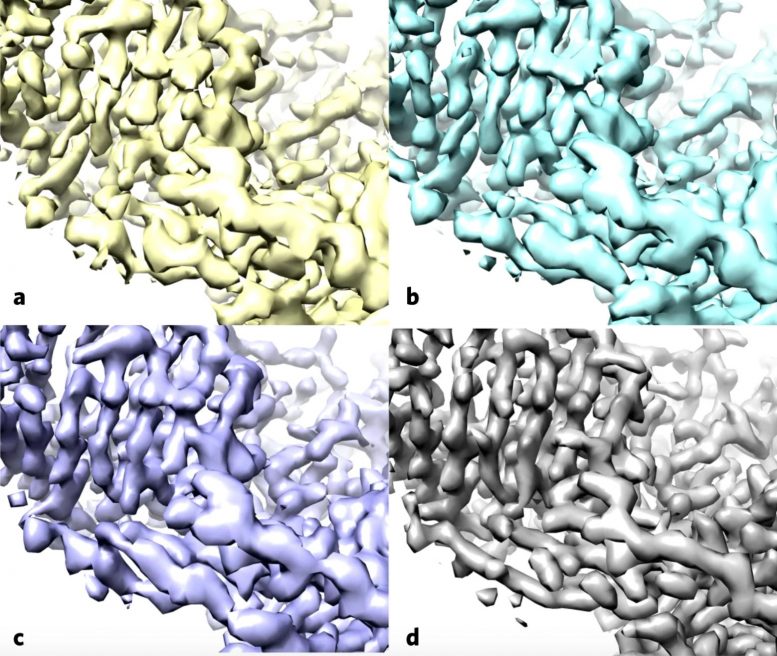
Using the enzyme β-galactosidase, also called lactase, as a test case, the researchers applied the standard methods (a) and then applied the improvement algorithm without (b) and with a filter to improve the uniformity of noise in the map (c), both of these maps are more similar to the deposited high-resolution protein structure map (d). Credit: Terwilliger et al./Nature Methods
“In biology, we gain so much by knowing a molecule’s structure,” said study co-author Paul Adams, Director of the Molecular Biophysics & Integrated Bioimaging Division at Berkeley Lab. “The improvements we see with this algorithm will make it easier for researchers to determine atomistic structural models from electron cryo-microscopy data. This is particularly consequential for modeling very important biological molecules, such as those involved in transcribing and translating the genetic code, which are often only seen in lower-resolution maps due to their large and complex multi-unit structures.”
The algorithm sharpens molecular maps by filtering the data based on existing knowledge of what molecules look like and how to best estimate and remove noise (unwanted and irrelevant data) in microscopy data. An approach with the same theoretical basis was previously used to improve structure maps generated from X-ray crystallography, and scientists have proposed its use in cryo-EM before. But, according to Adams, no one had been able to show definitive evidence that it worked for cryo-EM until now.
The team — composed of scientists from New Mexico Consortium, Los Alamos National Laboratory, Baylor College of Medicine, Cambridge University, and Berkeley Lab — first applied the algorithm to a publicly available map of the human protein apoferritin that is known to have 3.1-angstrom resolution (an angstrom is equal to a 10-billionth of a meter; for reference, the diameter of a carbon atom is estimated to be 2 angstroms). Then, they compared their enhanced version to another publicly available apoferritin reference map with 1.8-angstrom resolution, and found improved correlation between the two.
Next, the team used their approach on 104 map datasets from the Electron Microscopy Data Bank. For a large proportion of these map sets, the algorithm improved the correlation between the experimental map and the known atomic structure, and increased the visibility of details.
The authors note that the clear benefits of the algorithm in revealing important details in the data, combined with its ease of use — it is an automated analysis that can be performed on a laptop processor — will likely make it part of a standard part of the cryo-EM workflow moving forward. In fact, Adams has already added the algorithm’s source code to the Phenix software suite, a popular package for automated macromolecular structure solution for which he leads the development team.
This research was part of Berkeley Lab’s continued efforts to advance the capabilities of cryo-EM technology and to pioneer its use for basic science discoveries. Many of the breakthrough inventions that enabled the development of cryo-EM and later pushed it to its exceptional current resolution have involved Berkeley Lab scientists.
Reference: “Improvement of cryo-EM maps by density modification” by Thomas C. Terwilliger, Steven J. Ludtke, Randy J. Read, Paul D. Adams and Pavel V. Afonine, 17 August 2020, Nature Methods.
DOI: 10.1038/s41592-020-0914-9






Be the first to comment on "New Algorithm Sharpens Focus of World’s Most Powerful Microscopes – Improves Cryo-EM 3D Molecular Structure Maps"