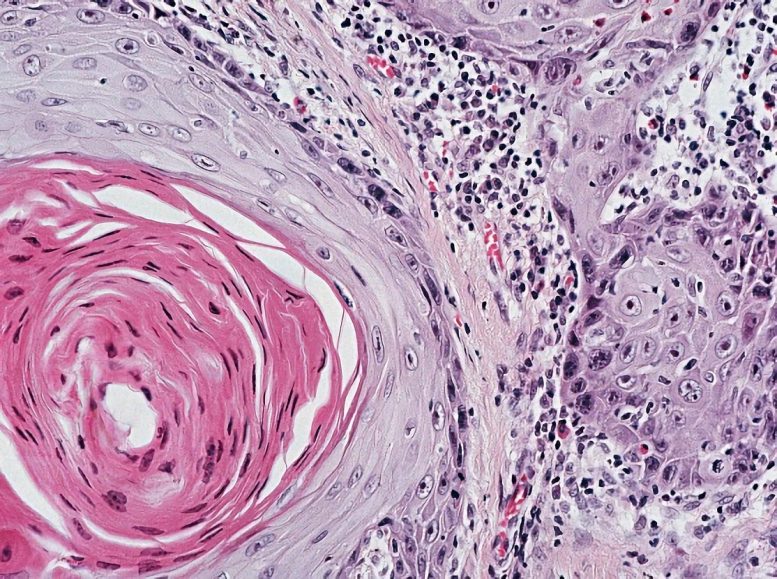
Squamous cell lung cancer. Credit: The Cancer Genome Atlas Research Network
A newly published study shows that squamous cell carcinoma is a cancer with diverse genomic causes, many of which are potentially susceptible to drug inhibition, and provides many new therapeutic opportunities for squamous cell carcinoma that would be suitable for clinical trials.
Squamous cell lung cancer kills more people each year than breast, colorectal or prostate cancer, ranking second only to lung adenocarcinoma in the number of deaths it causes. But unlike adenocarcinoma, squamous cell carcinoma has no treatments aimed at the specific genetic alterations that drive it. This may be about to change.
A new paper published online in Nature holds out hope that people with the second most common type of lung cancer may one day benefit from targeted therapies that have transformed treatments for patients with other types of lung cancer.
The Cancer Genome Atlas (TCGA) Research Network, led in part by scientists at the Broad Institute of Harvard and MIT, Dana-Farber Cancer Institute and Harvard Medical School, has identified many potential therapeutic targets based on the large number and variety of DNA alterations they discovered in most of the tumors they studied.
“This study clearly shows that squamous cell carcinoma, like lung adenocarcinoma, is a cancer with diverse genomic causes, many of which are potentially susceptible to drug inhibition,” said Matthew Meyerson, co-leader of the project within TCGA, Broad senior associate member, and HMS professor of pathology at Dana-Farber. “This provides many new therapeutic opportunities for squamous cell carcinoma that would be suitable for clinical trials.”
The first targeted treatments for lung adenocarcinoma, erlotinib (marketed as Tarceva) and gefitinib (marketed as Iressa), were aimed at mutations in the EGFR gene. But these drugs, like several being tested in clinical trials that target other genes altered in lung adenocarcinoma, do not help patients with squamous cell carcinoma. The TCGA effort, a multicenter consortium funded by the National Institutes of Health, is the first comprehensive genomic characterization of this lung cancer subtype. Squamous cell carcinoma causes approximately 400,000 deaths per year worldwide.
As in the three previous TCGA reports on glioblastoma, a type of brain cancer, on ovarian cancer and on colorectal cancer, the scientists used multiple large-scale approaches to highlight key molecular defects. They sequenced the protein-coding segments of the genomes of 178 squamous cell carcinoma tumors, along with normal tissue from the same patients. The researchers also sequenced the entire genomes of 19 tumor- and normal-tissue pairs. They mapped a diverse catalog of genomic alterations, including the rearrangements of chromosomes and other structural changes in regions of the genome that might not encode proteins but could control nearby genes involved in cancer development.
The comprehensive study confirmed some previously identified genomic alterations. For example, the TP53 gene was altered in 90 percent of tumors and the CDKN2A gene was inactivated in 72 percent of tumors. When these genes are switched off, tumors can grow unimpeded. CDKN2A may be susceptible to a kinase inhibitor, presenting an opportunity for clinical trials.
Overall, the researchers identified mutations or amplifications in three families of tyrosine kinases, which are enzymes that act like power switches for many cellular functions. Frequently altered in cancer, they are already being investigated as therapeutic targets in other cancers. The researchers also found genomic alterations in signaling pathways that could present important opportunities for treatment.
In another striking finding, the researchers discovered mutations in the HLA-A gene that hampered its function in tumors. These genes direct the arm of the immune system that discriminates between the body’s own tissues and foreign invaders. This is the first cancer in which these mutations have been found, but they are likely to occur in other cancers, Meyerson said.
“To our knowledge, this is the first example of a tumor that has a genomic mechanism for evading an immune response,” he said. “This may be important in understanding the immune response to squamous cell carcinoma and also in envisioning how immune-regulatory therapy might be used for this disease.”
While much work needs to be done, the scientists see many opportunities.
“When we see lung cancer patients, it’s almost a double standard. If you have lung adenocarcinoma, we can offer you molecular testing, we can put you in trials, we can put you on some targeted drugs,” said Peter Hammerman, a co-chair of the paper’s writing and analysis committee, an associate researcher at the Broad, a member of the thoracic oncology program and an HMS instructor in medicine at Dana-Farber. “If you have squamous cell lung cancer, you get the same treatment today you got 10 years ago, which is no more effective than it was 10 years ago. We’re just starting to see the first glimmers of hope in squamous cell lung cancer. This paper takes us to the next level in terms of identifying a number of potentially interesting targets to work on.”
The Broad’s participation in the TCGA network included leadership from three centers: the Genome Sequencing Center, with Broad founding director Eric Lander as principal investigator; one of six Genome Characterization Centers, led by Meyerson; and one of six Genome Data Analysis Centers, led by Gad Getz, director of Cancer Genome Computational Analysis at the Broad, and Broad senior associate member Lynda Chin, formerly at Dana-Farber and now at The University of Texas MD Anderson Cancer Center.
“I think it’s the hope of all of us who worked on this study that we’ll catalyze a large new set of clinical trials in squamous cell carcinoma,” Meyerson said.
Reference: “Comprehensive genomic characterization of squamous cell lung cancers” by The Cancer Genome Atlas Research Network, 9 September 2012, Nature.
DOI: 10.1038/nature11404
Other researchers from the Broad, Dana-Farber, Harvard, and their affiliates who contributed to the paper include Michael S. Lawrence, Douglas Voet, Rui Jing, Kristian Cibulskis, Andrey Sivachenko, Petar Stojanov, Aaron McKenna, Stacey Gabriel, Carrie Sougnez, Marcin Imielinski, Elena Helman, Bryan Hernandez, Nam H. Pho, Gordon Saksena, Andrew D. Cherniack, Steven E. Schumacher, Barbara Tabak, Scott L. Carter, Huy Nguyen, Robert C. Onofrio, Andrew Crenshaw, Kristin Ardlie, Rameen Beroukhim, Wendy Winckler, Michael Noble, Nils Gehlenborg, Alexei Protopopov, Angela Hadjipanayis, Semin Lee, Ruibin Xi, Lixing Yang, Xiaojia Ren, Sachet Shukla, Peng-Chieh Chen, Psalm Haseley, Eunjung Lee, Raju Kucherlapati, Daniel DiCara, Jinhua Zhang, Hailei Zhang, Chang-Jiun Wu, Yingchun Liu, Lihua Zou, Pei Lin, Juok Cho, Marc-Danie Nazaire, Jim Robinson, Helga Thorvaldsdottir, Jill Mesirov, Peter J. Park, Bruce Johnson, and David Kwiatkowski.





Be the first to comment on "Cancer Genome Atlas Finds Potential Drug Targets for Squamous Cell Lung Cancers"