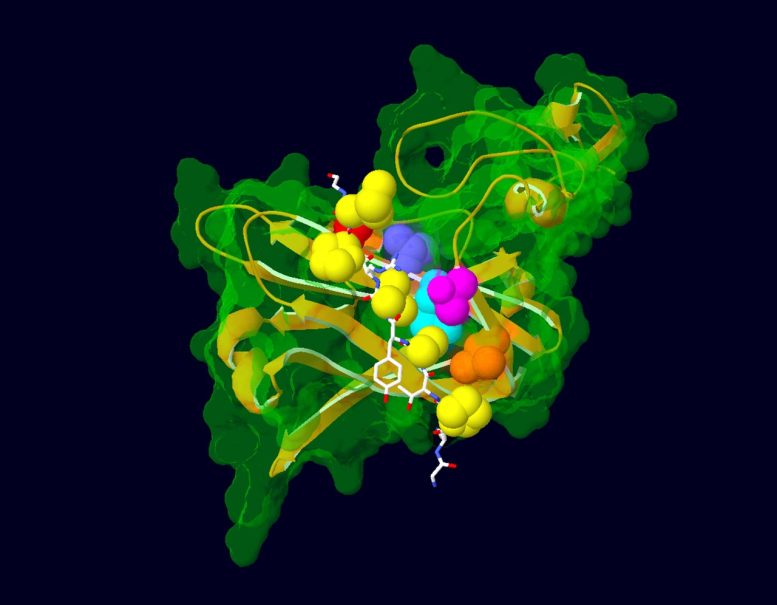
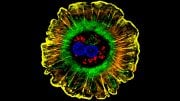
Model showing the interaction between a portion of the AFF3 protein (in white) and ubiquitin ligase (in green and gold), the protein that regulates its degradation. Amino acids mutated in KINSSHIP syndrome patients are shown as yellow atoms. The ubiquitin ligase amino acids with which they interact are depicted as colored atoms. Credit: Nicolas Guex © UNIL
New research reveals that both the excess and the deficiency of a single protein can lead to severe intellectual deficiencies. The discovery offers critical insights for early diagnosis of a rare developmental disorder.
A team of scientists presents a major step forward in the detection of a rare genetic disease. For the first time, the researchers show that both the accumulation and the deficiency of the so-called AFF3 protein are detrimental to development. The research was led by Alexandre Reymond, an expert in human genetics at the Center for Integrative Genomics (CIG) and professor at the Faculty of Biology and Medicine (FBM) of the University of Lausanne (UNIL).
The research, published today (May 30) in Genome Medicine, follows on from the group’s 2021 discovery of the KINSSHIP syndrome, caused by mutations in the AFF3 gene and resulting in intellectual disability, an increased risk for epilepsy, kidney malformations, and bone deformation in affected children.
Discovery of the genetic cause of KINSSHIP syndrome
KINSSHIP syndrome affects about thirty individuals worldwide. As a result, there are few documented cases and understanding of the disease remains limited, making early and accurate diagnosis challenging.
“In our previous study we demonstrated that this pathology resulted from an abnormal accumulation of the AFF3 protein. Meanwhile, available genetic data from individuals of the general population suggested that a lack of this same protein could be similarly deleterious,” explains Dr. Sissy Bassani, a postdoctoral researcher in Professor Reymond’s team and the lead author of the current study.
Large genome database points researchers to a new hypothesis
The geneticists formulated their hypothesis using gnomAD, a database containing genome sequences from several hundred thousand unrelated individuals. By mining the available data for AFF3 variants, the scientists found that loss-of-function mutations in this gene are rare, indicating their likely harmful nature. This implies that this gene plays a critical role and that its loss likely has detrimental consequences for the organism.
To test their hypothesis, the authors searched for individuals with only one copy of the gene, instead of the two normally present in the human genome. Collaborating with researchers from nine different countries across Europe and North America, they identified 21 patients with such an anomaly. They all showed similar but less severe symptoms than those of KINSSHIP syndrome patients.
Experiments reveal the developmental impact of AFF3 gene mutations
To demonstrate that both insufficient and excessive amounts of AFF3 are detrimental, the researchers used several different experimental systems: cells of patients, mice, and zebrafish. Artificially decreasing or increasing the protein quantity in zebrafish eggs revealed major developmental defects in the resulting fish embryos.
“These results confirm that a precise amount of AFF3 is crucial for proper embryonic development and that mutations affecting its function and/or dosage cause severe malformations,” concludes Prof. Reymond.
Impact for prenatal diagnostics
The authors’ findings are an important advancement for the diagnosis of this rare disorder, as testing for AAF3 mutations during fetal development could improve early detection of these gene defects.
Reference: “Variant-specific pathophysiological mechanisms of AFF3 differently influence transcriptome profiles” by Sissy Bassani, Jacqueline Chrast, Giovanna Ambrosini, Norine Voisin, Frédéric Schütz, Alfredo Brusco, Fabio Sirchia, Lydia Turban, Susanna Schubert, Rami Abou Jamra, Jan-Ulrich Schlump, Desiree DeMille, Pinar Bayrak-Toydemir, Gary Rex Nelson, Kristen Nicole Wong, Laura Duncan, Mackenzie Mosera, Christian Gilissen, Lisenka E. L. M. Vissers, Rolph Pfundt, Rogier Kersseboom, Hilde Yttervik, Geir Åsmund Myge Hansen, Marie Falkenberg Smeland, Kameryn M. Butler, Michael J. Lyons, Claudia M. B. Carvalho, Chaofan Zhang, James R. Lupski, Lorraine Potocki, Leticia Flores-Gallegos, Rodrigo Morales-Toquero, Florence Petit, Binnaz Yalcin, Annabelle Tuttle, Houda Zghal Elloumi, Lane McCormick, Mary Kukolich, Oliver Klaas, Judit Horvath, Marcello Scala, Michele Iacomino, Francesca Operto, Federico Zara, Karin Writzl, Aleš Maver, Maria K. Haanpää, Pia Pohjola, Harri Arikka, Anneke J. A. Kievit, Camilla Calandrini, Christian Iseli, Nicolas Guex and Alexandre Reymond, 30 May 2024, Genome Medicine.
DOI: 10.1186/s13073-024-01339-y





Be the first to comment on "Developmental Disorders: Researchers Link Protein Levels to Severe Intellectual Deficiencies"