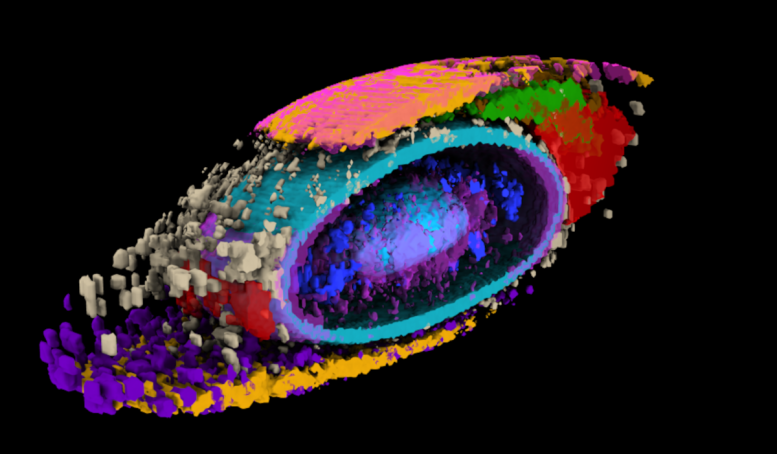
Integrated average morphed cell showing 17 select structures. Credit: Allen Institute for Cell Science
Researchers unveil a new method to visualize cell organization.
Working with hundreds of thousands of high-resolution images, researchers from the Allen Institute for Cell Science, a division of the Allen Institute, put numbers on the internal organization of human cells — a biological concept that has proven incredibly difficult to quantify until now.
The scientists also documented the diverse cell shapes of genetically identical cells grown under similar conditions in their work. Their findings were recently published in the journal Nature.
“The way cells are organized tells us something about their behavior and identity,” said Susanne Rafelski, Ph.D., Deputy Director of the Allen Institute for Cell Science, who led the study along with Senior Scientist Matheus Viana, Ph.D. “What’s been missing from the field, as we all try to understand how cells change in health and disease, is a rigorous way to deal with this kind of organization. We haven’t yet tapped into that information.”
This study provides a roadmap for biologists to understand the organization of different kinds of cells in a measurable, quantitative way, Rafelski said. It also reveals some key organizational principles of the cells the Allen Institute team studies, which are known as human induced pluripotent stem cells.
Understanding how cells organize themselves under healthy conditions — and the full range of variability contained within “normal” — can help scientists better understand what goes wrong in disease. The image dataset, genetically engineered stem cells, and code that went into this study are all publicly available for other scientists in the community to use.
“Part of what makes cell biology seem intractable is the fact that every cell looks different, even when they are the same type of cell. This study from the Allen Institute shows that this same variability that has long plagued the field is, in fact, an opportunity to study the rules by which a cell is put together,” said Wallace Marshall, Ph.D., Professor of Biochemistry and Biophysics at the University of California, San Francisco, and a member of the Allen Institute for Cell Science’s Scientific Advisory Board. “This approach is generalizable to virtually any cell, and I expect that many others will adopt the same methodology.”
Computing the pear-ness of our cells
In a body of work launched more than seven years ago, the Allen Institute team first built a collection of stem cells genetically engineered to light up different internal structures under a fluorescent microscope. With cell lines in hand that label 25 individual structures, the scientists then captured high-resolution, 3D images of more than 200,000 different cells.
All this to ask one seemingly straightforward question: How do our cells organize their interiors?
Getting to the answer, it turned out, is really complex. Imagine setting up your office with hundreds of different pieces of furniture, all of which need to be readily accessed, and many of which need to move freely or interact depending on their task. Now imagine your office is a sac of liquid surrounded by a thin membrane, and many of those hundreds of pieces of furniture are even smaller bags of liquid. Talk about an interior design nightmare.
The scientists wanted to know: How do all those tiny cellular structures arrange themselves compared to each other? Is “structure A” always in the same place, or is it random?
The team ran into a challenge comparing the same structure between two different cells. Even though the cells under study were genetically identical and reared in the same laboratory environment, their shapes varied substantially. The scientists realized that it would be impossible to compare the position of structure A in two different cells if one cell was short and blobby and the other was long and pear-shaped. So they put numbers on those stubby blobs and elongated pears.
Using computational analyses, the team developed what they call a “shape space” that objectively describes each stem cell’s external shape. That shape space includes eight different dimensions of shape variation, things like height, volume, elongation, and the aptly described “pear-ness” and “bean-ness.” The scientists could then compare apples to apples (or beans to beans), looking at the organization of cellular structures inside all similarly shaped cells.
“We know that in biology, shape, and function are interrelated, and understanding cell shape is important to understand how the cells function,” Viana said. “We’ve come up with a framework that allows us to measure a cell’s shape, and the moment you do that you can find cells that are similar shapes, and for those cells, you can then look inside and see how everything is arranged.”
Strict organization
When they looked at the position of the 25 highlighted structures, comparing those structures in groups of cells with similar shapes, they found that all the cells set up shop in remarkably similar ways. Despite the massive variations in cell shape, their internal organization was strikingly consistent.
If you’re looking at how thousands of white-collar workers arrange their furniture in a high-rise office building, it’s as if every worker put their desk smack in the middle of their office and their filing cabinet precisely in the far-left corner, no matter the size or shape of the office. Now say you found one office with a filing cabinet thrown on the floor and papers strewn everywhere — that might tell you something about the state of that particular office and its occupant.
The same goes for cells. Finding deviations from the normal state of affairs could give scientists important information about how cells change when they transition from stationary to mobile, are getting ready to divide, or about what goes wrong at the microscopic level in disease. The researchers looked at two variations in their dataset — cells at the edges of colonies of cells, and cells that were undergoing division to create new daughter cells, a process known as mitosis. In these two states, the scientists were able to find changes in internal organization correlating to the cells’ different environments or activities.
“This study brings together everything we’ve been doing at the Allen Institute for Cell Science since the institute was launched,” said Ru Gunawardane, Ph.D., Executive Director of the Allen Institute for Cell Science. “We built all of this from scratch, including the metrics to measure and compare different aspects of how cells are organized. What I’m truly excited about is how we and others in the community can now build on this and ask questions about cell biology that we could never ask before.”
Reference: “Integrated intracellular organization and its variations in human iPS cells” by Matheus P. Viana, Jianxu Chen, Theo A. Knijnenburg, Ritvik Vasan, Calysta Yan, Joy E. Arakaki, Matte Bailey, Ben Berry, Antoine Borensztejn, Eva M. Brown, Sara Carlson, Julie A. Cass, Basudev Chaudhuri, Kimberly R. Cordes Metzler, Mackenzie E. Coston, Zach J. Crabtree, Steve Davidson, Colette M. DeLizo, Shailja Dhaka, Stephanie Q. Dinh, Thao P. Do, Justin Domingus, Rory M. Donovan-Maiye, Alexandra J. Ferrante, Tyler J. Foster, Christopher L. Frick, Griffin Fujioka, Margaret A. Fuqua, Jamie L. Gehring, Kaytlyn A. Gerbin, Tanya Grancharova, Benjamin W. Gregor, Lisa J. Harrylock, Amanda Haupt, Melissa C. Hendershott, Caroline Hookway, Alan R. Horwitz, H. Christopher Hughes, Eric J. Isaac, Gregory R. Johnson, Brian Kim, Andrew N. Leonard, Winnie W. Leung, Jordan J. Lucas, Susan A. Ludmann, Blair M. Lyons, Haseeb Malik, Ryan McGregor, Gabe E. Medrash, Sean L. Meharry, Kevin Mitcham, Irina A. Mueller, Timothy L. Murphy-Stevens, Aditya Nath, Angelique M. Nelson, Sandra A. Oluoch, Luana Paleologu, T. Alexander Popiel, Megan M. Riel-Mehan, Brock Roberts, Lisa M. Schaefbauer, Magdalena Schwarzl, Jamie Sherman, Sylvain Slaton, M. Filip Sluzewski, Jacqueline E. Smith, Youngmee Sul, Madison J. Swain-Bowden, W. Joyce Tang, Derek J. Thirstrup, Daniel M. Toloudis, Andrew P. Tucker, Veronica Valencia, Winfried Wiegraebe, Thushara Wijeratna, Ruian Yang, Rebecca J. Zaunbrecher, Ramon Lorenzo D. Labitigan, Adrian L. Sanborn, Graham T. Johnson, Ruwanthi N. Gunawardane, Nathalie Gaudreault, Julie A. Theriot and Susanne M. Rafelski, 4 January 2023, Nature.
DOI: 10.1038/s41586-022-05563-7





Still believe it’s all due to a bunch of random errors?