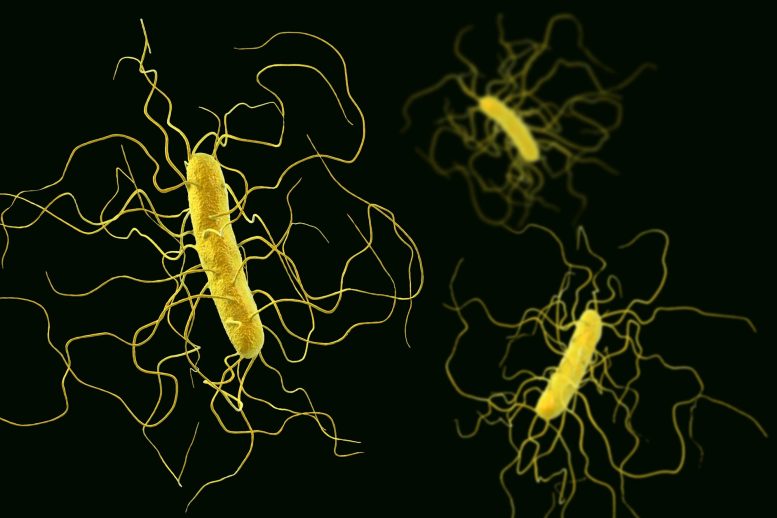
Illustration of Clostridioides difficile bacterium with peritrichous flagella. Scientists uncovered a dual mechanism in C. diff that bolsters its antibiotic resistance, which could pave the way for more effective treatment strategies against resistant bacteria.
A species of ordinary gut bacteria that we all carry flourishes when the intestinal flora is knocked out by a course of antibiotics. Since the bacteria is naturally resistant to many antibiotics, it causes problems, particularly in healthcare settings. A study led by Lund University in Sweden now shows how two molecular mechanisms can work together make the bacterium extra resistant. “Using this knowledge, we hope to be able to design even better medicines,” says Vasili Hauryliuk, senior lecturer at Lund University, who led the study.
The threat from antibiotic resistant bacteria is as well-known as it is grave. Last year, The Lancet reported that an estimated 1.27 million people died in 2019 as a result of bacterial infection that could not be treated with existing medicines. To tackle this threat, it is essential to understand the underpinning molecular mechanisms.
During antibiotic treatment, the normal intestinal flora is disturbed, which provides an opportunity for antibiotic resistant bacterial pathogens that are otherwise suppressed though competition with the “good” gut bacteria. One of the most problematic bacterial species is Clostridioides difficile, C. diff. It is found in our intestines, is resistant to antibiotic treatments and can cause serious diarrheal infections. The bacteria’s ability to create spores means it is easily spread and therefore causes problems in healthcare settings, resulting both in increased mortality and extended treatment times.
“Instead of the antibiotic saving you, in this case, it promotes a secondary bacterial infection,” says Vasili Hauryliuk.
“The risk of infection with C. diff is known to increase after treatment with an antibiotic called clindamycin, but the reason for this was unknown. Our research showed a novel protein conveys resistance to the class of antibiotics to which clindamycin belongs,” says Obana Nozomu, assistant professor at the University of Tsukuba and one of the researchers behind the study.
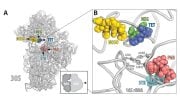
The mechanism of C. diff resistance has been investigated in an international collaboration between researchers in Sweden, Japan, the United Kingdom, USA, Estonia, and Germany, and the results of this study have been published in Nucleic Acids Research. When researchers have identified a novel protein that is responsible for the resistance. The protein works on the ribosome – the molecular factory that produces the proteins in the bacteria, giving the bacteria its abilities. The ribosome is one of the primary antibiotic targets: if proteins cannot be synthesized, the bacteria will not grow, replicate and cause the infection.
“This newly discovered protein kicks the antibiotic molecule out of the ribosome. We also saw that it combines with another resistance factor. The second chemically modifies the ribosome so that the antibiotic molecules to bind less tightly to it. The extra-potent resistance is the result of two mechanisms, two factors, which combine and in so doing give the bacteria its ‘superpowers’ against antibiotics,” says Gemma C. Atkinson, senior lecturer at Lund University and co-author of the article.
The researchers used cryogenic electron microscopy in order to study the resistance mechanisms against antibiotics on a molecular level. This knowledge opens the way for new treatment strategies against resistance and the infections that the bacteria cause.
“A couple of years ago, Andrew G. Myers lab at Harvard University has developed a new generation of ribosome-binding antibiotics, known as iboxamycin. It is a very potent medicine that knocks out ‘ordinary’ C. diff bacteria. The results of this study, however, show that C. diff strains that have both resistance factors are, unfortunately, resistant to this antibiotic as well. This means that it is necessary to design antibiotic molecules that bind even tighter in order to overpower this kind of resistance. We now collaborate with the Myers group on this direction.” says Vasili Hauryliuk.
This study also found that certain antibiotics that target the ribosome induce the production of the resistance factor. This may also provide clues for designing new antibiotic molecules, since resistance cannot be induced if resistance factors are not synthesized.
Reference: “Genome-encoded ABCF factors implicated in intrinsic antibiotic resistance in Gram-positive bacteria: VmlR2, Ard1 and CplR” by Nozomu Obana, Hiraku Takada, Caillan Crowe-McAuliffe, Mizuki Iwamoto, Artyom A Egorov, Kelvin J Y Wu, Shinobu Chiba, Victoriia Murina, Helge Paternoga, Ben I C Tresco, Nobuhiko Nomura, Andrew G Myers, Gemma C Atkinson, Daniel N Wilson and Vasili Hauryliuk, 23 March 2023, Nucleic Acids Research.
DOI: 10.1093/nar/gkad193
The study was carried out with funding from the Swedish Research Council, the Swedish Cancer Society, the Knut and Alice Wallenberg Foundation, the European Research Council, National Institutes of Health and the research councils of Estonia and Germany.


So they know WHY you’re going to die a horrible death, but they can’t do a darn thing about it. Yeah, in other words, not very helpful. It’s like knowing how much energy is required to build a wormhole, but realizing you’d have to turn Jupiter into a giant anti-matter reaction to generate it. Oops.