
An international research consortium co-led by scientists from multiple universities has released a series of studies detailing new high-quality reference genomes from 50 primate species, 27 of which were sequenced for the first time. The findings offer fresh insights into primate evolution, speciation, genomic diversity, and the evolution of brain and other traits, enhancing our understanding of the human genetic architecture, primate diversification, and significant evolutionary phenomena like hybridization and incomplete lineage sorting.
Analyses of 50 primate genomes through comparative genomics unveil essential genetic processes involved in primate speciation, adaptive phenotypic changes, and the evolution of social systems.
A series of publications from the first phase program of the Primate Genome Consortium presented high-quality reference genomes from 50 primate species, including 27 that were sequenced for the first time.
The studies provide new insight into the process of speciation, genomic diversity, social evolution, the evolution of sex chromosomes, the brain, and other biological traits.
The research was co-led by Guojie Zhang from the Centre for Evolutionary & Organismal Biology at Zhejiang University, Dong-Dong Wu at the Kunming Institute of Zoology, Xiao-Guang Qi at Northwest University, Li Yu at Yunnan University, Mikkel Heide Schierup at Aarhus University, and Yang Zhou at BGI-Research.
Large-scale phylogenomic studies reveal the genetic mechanisms underlying the evolutionary history and phenotypic innovations in primates
The comparative analysis of primate genomes within a phylogenetic context is crucial for understanding the evolution of the human genetic architecture and the inter-species genomic differences associated with primate diversification. Previous studies of primate genomes have focused mainly on primate species closely related to humans and were constrained by the lack of broader phylogenetic coverage.
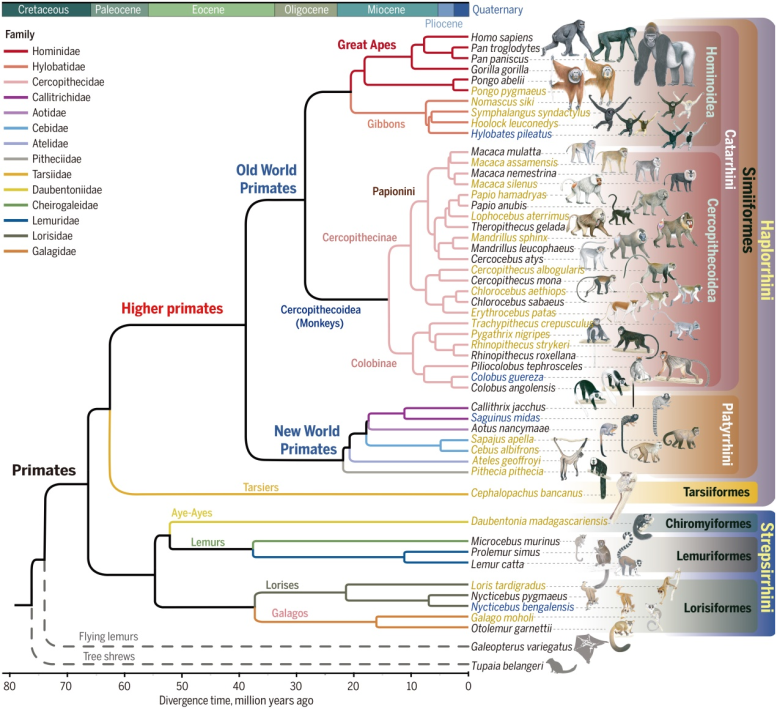
Genomic phylogeny of primates. Credit: Dong-Dong Wu.
“Although there are more than 500 primate species worldwide, currently, only 23 representative non-human primates species have had their genomes published, leaving 72% of genera remain unsequenced, which creates significant knowledge gaps in understanding their evolutionary history,” Dong-Dong Wu states.
To address this gap, they performed high-quality genome sequencing using long-read sequencing technologies on 27 primate species, including basal lineages that had not been fully sequenced before. Combining this with previously published primate genomes, the project conducted phylogenomic studies of 50 primate species representing 38 genera and 14 families to gain new insights into their genomic and phenotypic evolution.
“Based on full genome data, we have generated a highly resolved phylogeny and estimated the emergence of crown Primates between 64.95 and 68.29 million years ago overlapping the Cretaceous/Tertiary boundary,” Dong-Dong Wu states.
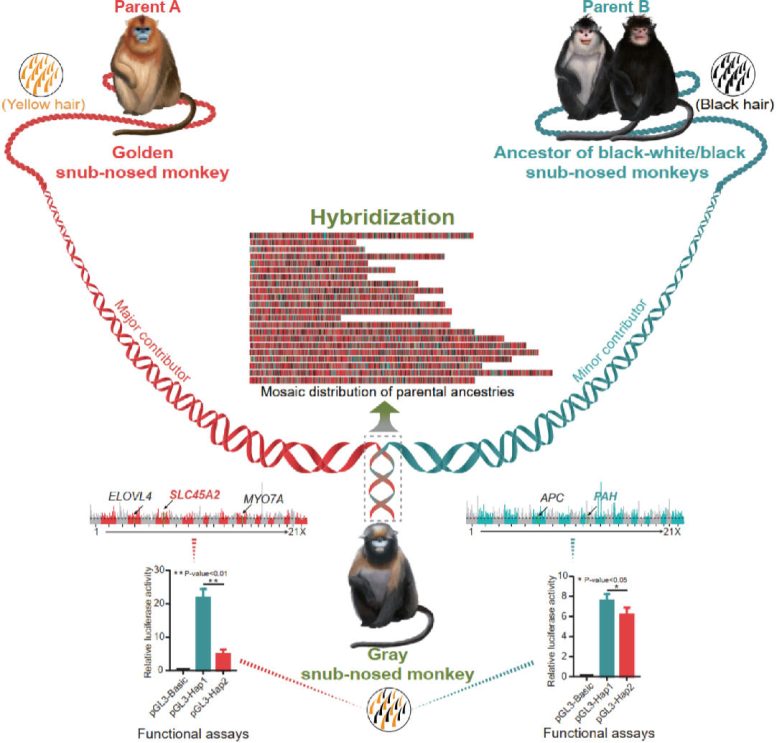
The hybrid origin of gray snub-nosed monkey. Credit: Li Yu
The study reported detailed genomic rearrangements across primate lineages and identified thousands of candidate genes that have undergone adaptive natural selection at different ancestral branches of the phylogeny. This includes genes that are important for the development of the nervous, skeletal, digestive, and sensory systems, all of which are likely to have contributed to evolutionary innovations and adaptations of primates.
“It is surprising to see that so many genomic changes involving brain-related genes occurred in the common ancestor of the Simian group which includes New-world monkey, Old-world monkey, and great apes,” states Guojie Zhang, “These genomic innovations evolving deep in time at this ancestral node might have paved the way for the further evolution of human unique traits.”
Pervasive incomplete lineage sorting illuminates speciation and selection in primates
Although it has been well-recognized that chimpanzees and bonobos are the most closely related species to humans, 15% of our genome is closer to another great ape, the gorilla. This is primarily due to the special evolutionary event called incomplete-lineage sorting (ILS), where the ancestral genetic polymorphism randomly sorts into the descendent species. The study investigated the speciation events during the primate evolution and found ILS occurred frequently in all 29 major ancestral nodes across primates with some nodes having over 50% of the genome affected by ILS.
“The genetic diversification process does not follow a bifurcation tree-like topology as we normally know for speciation process, it is more like a complicated net,” Guojie Zhang said. ”It is important to investigate the evolutionary process of each individual gene, which could also affect the evolution of phenotypes across species.”

Gray snub-nosed monkey (Rhinopithecus brelichi). Credit: Gui-Yun Li
Incomplete lineage sorting (ILS) exhibits extensive variation along the genome, primarily driven by recombination. “We observed that ILS is reduced more on the X chromosome than autosomes compared to what would be expected under neutral evolution, suggesting a higher impact of natural selection on the X chromosome during primate evolution,” Mikkel Heide Schierup states.
The study exploits ILS to perform molecular dating of speciation events solely based on genome data, without fossil calibration and found the new dating results were highly consistent with the dating with the fossil record. “This suggests that molecular dating provides an accurate estimate of speciation time even without the fossil records”, says the first author of this paper, Iker Rivas-González.
Hybridization into species events
Hybridization is increasingly recognized as an important evolutionary force for generating species and phenotypic diversity in plants and animals. This is especially common in lineages that can tolerate whole genome duplication and increased levels of ploidy. However, speciation by hybridization has been rarely reported in mammals.
Utilizing full genome data, the team discovered that the gray snub-nosed monkey Rhinopithecus brelichi was a descendent species from the hybridization between the morphologically differentiated species, the golden snub-nosed monkey R. roxellana and the common ancestor of black-white snub-nosed monkey R. bieti and the black snub-nosed monkey R. strykeri.
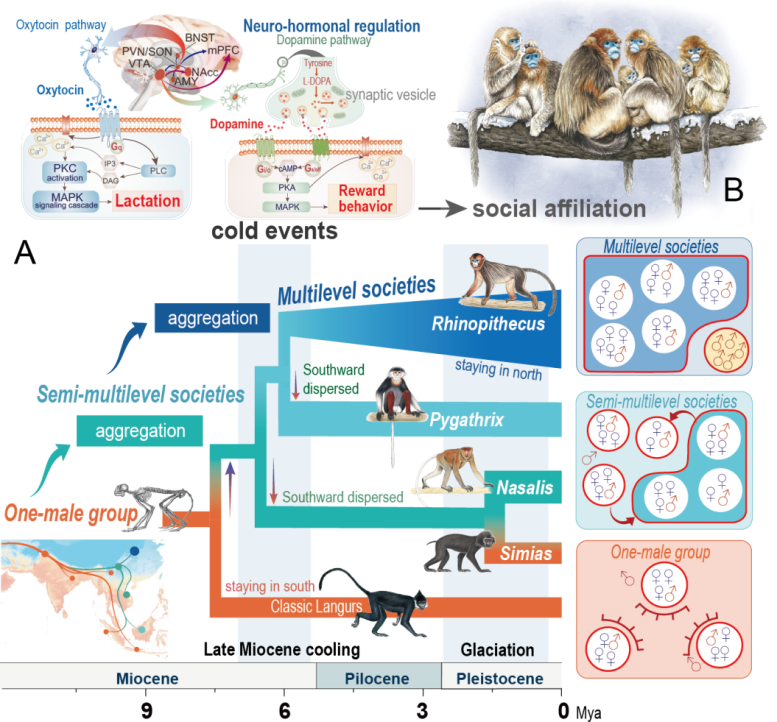
Cold promotes the social evolution of the Asian langurs. Credit: Xiao-Guang Qi.
“To our knowledge, this is the first time that a hybrid speciation event is recorded in primates,” stated Li Yu.
This study further identifies key genes in R. brelichi that derived from each parental lineage which may have contributed to the mosaic coat coloration in this species and likely promoted premating reproductive isolation of the hybrid species from the parental lineage.
Multidisciplinary intersection reveals the genetic mechanisms of social complexity in Asian langurs
Primates have very diverse social systems, however, the biological mechanisms underlying social evolution remain poorly known. The classical socioecological model hypothesized that the diversity of social systems evolved as a response to environmental changes.
The study used Asian colobine monkeys as a model system, as this group of species underwent a staged social evolution process from a one-male, multi-female unit to complex multi-level social forms. They have re-constructed the speciation process of this group using the full genome data and found a strong correlation between the environmental temperature and the group size of the species.
The primate species living in colder environments tend to live in larger groups. The ancient ice ages drove the social evolution of these primates, promoting the aggregation of spreading northern odd-nosed monkey species into nested multi-level social forms.
During this transition, odd-nosed monkeys exhibited positive selection in many genes related to cold adaptation and the nervous system. “The snub-nosed monkeys seem to have a longer mother-infant bond, which probably increased infant survival in cold environments, The DA/OXT receptors are important neurohormones in mediating social bonding. This signal pathway has been enhanced in odd-nosed monkeys and promoted the social affiliation, cohesion, and cooperation among adults of this species,” Xiao-Guang Qi states.
References: “Phylogenomic analyses provide insights into primate evolution” by Yong Shao, Long Zhou, Fang Li, Lan Zhao, Bao-Lin Zhang, Feng Shao, Jia-Wei Chen, Chun-Yan Chen, Xupeng Bi, Xiao-Lin Zhuang, Hong-Liang Zhu, Jiang Hu, Zongyi Sun, Xin Li, Depeng Wang, Iker Rivas-González, Sheng Wang, Yun-Mei Wang, Wu Chen, Gang Li, Hui-Meng Lu, Yang Liu, Lukas F. K. Kuderna, Kyle Kai-How Farh, Peng-Fei Fan, Li Yu, Ming Li, Zhi-Jin Liu, George P. Tiley, Anne D. Yoder, Christian Roos, Takashi Hayakawa, Tomas Marques-Bonet, Jeffrey Rogers, Peter D. Stenson, David N. Cooper, Mikkel Heide Schierup, Yong-Gang Yao, Ya-Ping Zhang, Wen Wang, Xiao-Guang Qi, Guojie Zhang and Dong-Dong Wu, 1 June 2023, Science.
DOI: 10.1126/science.abn6919
“Pervasive incomplete lineage sorting illuminates speciation and selection in primates” by Iker Rivas-González, Marjolaine Rousselle, Fang Li, Long Zhou, Julien Y. Dutheil, Kasper Munch, Yong Shao, Dongdong Wu, Mikkel H. Schierup and Guojie Zhang, 2 June 2023, Science.
DOI: 10.1126/science.abn4409
“Hybrid origin of a primate, the gray snub-nosed monkey” by Hong Wu, Zefu Wang, Yuxing Zhang, Laurent Frantz, Christian Roos, David M. Irwin, Chenglin Zhang, Xuefeng Liu, Dongdong Wu, Song Huang, Tongtong Gu, Jianquan Liu and Li Yu, 2 June 2023, Science.
DOI: 10.1126/science.abl4997
“Adaptations to a cold climate promoted social evolution in Asian colobine primates” by Xiao-Guang Qi, Jinwei Wu, Lan Zhao, Lu Wang, Xuanmin Guang, Paul A. Garber, Christopher Opie, Yuan Yuan, Runjie Diao, Gang Li, Kun Wang, Ruliang Pan, Weihong Ji, Hailu Sun, Zhi-Pang Huang, Chunzhong Xu, Arief B. Witarto, Rui Jia, Chi Zhang, Cheng Deng, Qiang Qiu, Guojie Zhang, Cyril C. Grueter, Dongdong Wu and Baoguo Li, 2 June 2023, Science.
DOI: 10.1126/science.abl8621
“The landscape of tolerated genetic variation in humans and primates” by Hong Gao, Tobias Hamp, Jeffrey Ede, Joshua G. Schraiber, Jeremy McRae, Moriel Singer-Berk, Yanshen Yang, Anastasia S. D. Dietrich, Petko P. Fiziev, Lukas F. K. Kuderna, Laksshman Sundaram, Yibing Wu, Aashish Adhikari, Yair Field, Chen Chen, Serafim Batzoglou, Francois Aguet, Gabrielle Lemire, Rebecca Reimers, Daniel Balick, Mareike C. Janiak, Martin Kuhlwilm, Joseph D. Orkin, Shivakumara Manu, Alejandro Valenzuela, Juraj Bergman, Marjolaine Rousselle, Felipe Ennes Silva, Lidia Agueda, Julie Blanc, Marta Gut, Dorien de Vries, Ian Goodhead, R. Alan Harris, Muthuswamy Raveendran, Axel Jensen, Idriss S. Chuma, Julie E. Horvath, Christina Hvilsom, David Juan, Peter Frandsen, Fabiano R. de Melo, Fabrício Bertuol, Hazel Byrne, Iracilda Sampaio, Izeni Farias, João Valsecchi do Amaral, Mariluce Messias, Maria N. F. da Silva, Mihir Trivedi, Rogerio Rossi, Tomas Hrbek, Nicole Andriaholinirina, Clément J. Rabarivola, Alphonse Zaramody, Clifford J. Jolly, Jane Phillips-Conroy, Gregory Wilkerson, Christian Abee, Joe H. Simmons, Eduardo Fernandez-Duque, Sree Kanthaswamy, Fekadu Shiferaw, Dongdong Wu, Long Zhou, Yong Shao, Guojie Zhang, Julius D. Keyyu, Sascha Knauf, Minh D. Le, Esther Lizano, Stefan Merker, Arcadi Navarro, Thomas Bataillon, Tilo Nadler, Chiea Chuen Khor, Jessica Lee, Patrick Tan, Weng Khong Lim, Andrew C. Kitchener, Dietmar Zinner, Ivo Gut, Amanda Melin, Katerina Guschanski, Mikkel Heide Schierup, Robin M. D. Beck, Govindhaswamy Umapathy, Christian Roos, Jean P. Boubli, Monkol Lek, Shamil Sunyaev, Anne O’Donnell-Luria, Heidi L. Rehm, Jinbo Xu, Jeffrey Rogers, Tomas Marques-Bonet and Kyle Kai-How Farh, 2 June 2023, Science.
DOI: 10.1126/science.abn8197
“A global catalog of whole-genome diversity from 233 primate species” by Lukas F. K. Kuderna, Hong Gao, Mareike C. Janiak, Martin Kuhlwilm, Joseph D. Orkin, Thomas Bataillon, Shivakumara Manu, Alejandro Valenzuela, Juraj Bergman, Marjolaine Rousselle, Felipe Ennes Silva, Lidia Agueda, Julie Blanc, Marta Gut, Dorien de Vries, Ian Goodhead, R. Alan Harris, Muthuswamy Raveendran, Axel Jensen, Idrissa S. Chuma, Julie E. Horvath, Christina Hvilsom, David Juan, Peter Frandsen, Joshua G. Schraiber, Fabiano R. de Melo, Fabrício Bertuol, Hazel Byrne, Iracilda Sampaio, Izeni Farias, João Valsecchi, Malu Messias, Maria N. F. da Silva, Mihir Trivedi, Rogerio Rossi, Tomas Hrbek, Nicole Andriaholinirina, Clément J. Rabarivola, Alphonse Zaramody, Clifford J. Jolly, Jane Phillips-Conroy, Gregory Wilkerson, Christian Abee, Joe H. Simmons, Eduardo Fernandez-Duque, Sree Kanthaswamy, Fekadu Shiferaw, Dongdong Wu, Long Zhou, Yong Shao, Guojie Zhang, Julius D. Keyyu, Sascha Knauf, Minh D. Le, Esther Lizano, Stefan Merker, Arcadi Navarro, Tilo Nadler, Chiea Chuen Khor, Jessica Lee, Patrick Tan, Weng Khong Lim, Andrew C. Kitchener, Dietmar Zinner, Ivo Gut, Amanda D. Melin, Katerina Guschanski, Mikkel Heide Schierup, Robin M. D. Beck, Govindhaswamy Umapathy, Christian Roos, Jean P. Boubli, Jeffrey Rogers, Kyle Kai-How Farh and Tomas Marques Bonet, 1 June 2023, Science.
DOI: 10.1126/science.abn7829
“Genome-wide coancestry reveals details of ancient and recent male-driven reticulation in baboons” by Erik F. Sørensen, R. Alan Harris, Liye Zhang, Muthuswamy Raveendran, Lukas F. K. Kuderna, Jerilyn A. Walker, Jessica M. Storer, Martin Kuhlwilm, Claudia Fontsere, Lakshmi Seshadri, Christina M. Bergey, Andrew S. Burrell, Juraj Bergman, Jane E. Phillips-Conroy, Fekadu Shiferaw, Kenneth L. Chiou, Idrissa S. Chuma, Julius D. Keyyu, Julia Fischer, Marie-Claude Gingras, Sejal Salvi, Harshavardhan Doddapaneni, Mikkel H. Schierup, Mark A. Batzer, Clifford J. Jolly, Sascha Knauf, Dietmar Zinner, Kyle K.-H. Farh, Tomas Marques-Bonet, Kasper Munch, Christian Roos and Jeffrey Rogers, 2 June 2023, Science.
DOI: 10.1126/science.abn8153
“Rare penetrant mutations confer severe risk of common diseases” by Petko P. Fiziev, Jeremy McRae, Jacob C. Ulirsch, Jacqueline S. Dron, Tobias Hamp, Yanshen Yang, Pierrick Wainschtein, Zijian Ni, Joshua G. Schraiber, Hong Gao, Dylan Cable, Yair Field, Francois Aguet, Marc Fasnacht, Ahmed Metwally, Jeffrey Rogers, Tomas Marques-Bonet, Heidi L. Rehm, Anne O’Donnell-Luria, Amit V. Khera and Kyle Kai-How Farh, 2 June 2023, Science.
DOI: 10.1126/science.abo1131
“Comparative genomics reveals the hybrid origin of a macaque group” by Bao-Lin Zhang, Wu Chen, Zefu Wang, Wei Pang, Meng-Ting Luo, Sheng Wang, Yong Shao, Wen-Qiang He, Yuan Deng, Long Zhou, Jiawei Chen, Min-Min Yang, Yajiang Wu, Lu Wang, Hugo Fernández-Bellon, Sandra Molloy, Hélène Meunier, Fanélie Wanert, Lukas Kuderna, Tomas Marques-Bonet, Christian Roos, Xiao-Guang Qi, Ming Li, Zhijin Liu, Mikkel Heide Schierup, David N. Cooper, Jianquan Liu, Yong-Tang Zheng, Guojie Zhang and Dong-Dong Wu, 1 June 2023, Science Advances.
DOI: 10.1126/sciadv.add3580
“Lineage-specific accelerated sequences underlying primate evolution” by Xupeng Bi, Long Zhou, Jin-Jin Zhang, Shaohong Feng, Mei Hu, David N. Cooper, Jiangwei Lin, Jiali Li, Dong-Dong Wu and Guojie Zhang, 1 June 2023, Science Advances.
DOI: 10.1126/sciadv.adc9507
“Eighty million years of rapid evolution of the primate Y chromosome” by Yang Zhou, Xiaoyu Zhan, Jiazheng Jin, Long Zhou, Juraj Bergman, Xuemei Li, Marjolaine Marie C. Rousselle, Meritxell Riera Belles, Lan Zhao, Miaoquan Fang, Jiawei Chen, Qi Fang, Lukas Kuderna, Tomas Marques-Bonet, Haruka Kitayama, Takashi Hayakawa, Yong-Gang Yao, Huanming Yang, David N. Cooper, Xiaoguang Qi, Dong-Dong Wu, Mikkel Heide Schierup and Guojie Zhang, 2 June 2023, Nature Ecology & Evolution.
DOI: 10.1038/s41559-022-01974-x








Be the first to comment on "New Research Unlocks Hidden Secrets of Primate Evolution"