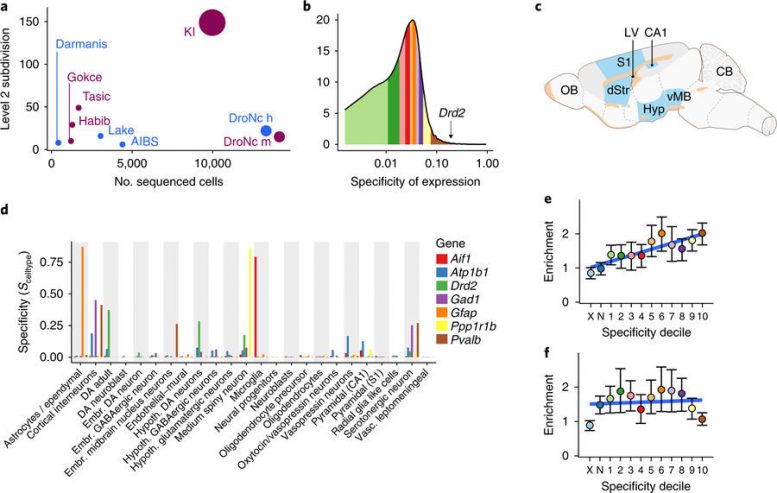
Specificity metric calculated from single-cell transcriptome sequencing data can be used to test for increased burden of schizophrenia-SNP heritability in brain cell types. a, Comparison of level 2 cell-type categories and number of cells subjected to snRNA-seq or scRNA-seq from brain tissue. Plum-colored circles are mouse studies, and blue circles are human studies. The number of different tissues is reflected in the size of the circle (see Supplementary Table 2 for citations). AIBS, Allen Institute for Brain Science; KI, Karolinska Institutet. b, Histogram of specificity metric (SMSN,KI) for MSNs from the KI superset level 1. Colored regions indicate deciles (the brown region contains the genes most specific to MSNs). Specificity value for dopamine receptor D2 (Drd2, SMSN,KI,Drd2 = 0.17) is indicated by the arrow. c, Schematic highlighting the brain regions sampled in the KI dataset in blue. d, Specificity values in the KI level 1 dataset for a range of known cell type markers. Embr., embryonic; Hypoth., hypothalamic. e, Enrichment of schizophrenia-SNP heritability in each of the specificity deciles for MSNs (calculated using LDSC). Color of dots corresponds to regions of the specificity matrix in b. The light blue dot (marked “X”) represents all SNPs that map onto named transcripts that are not MGI-annotated genes or that map onto a gene which does not have a 1:1 mouse:human ortholog. The dark blue dot (marked “N”) represents all SNPs that map onto genes not expressed in MSNs. Blue line shows the linear regression slope fitted to the enrichment values. f, Enrichment of height-SNP heritability in each of the specificity deciles for MSNs. Colors as in e. In e and f, error bars indicate the 95% confidence intervals. Nature Genetics (2018) doi:10.1038/s41588-018-0129-5
Scientists at Karolinska Institutet in Sweden and University of North Carolina, USA, have identified the cell types underlying schizophrenia in a new study published in Nature Genetics. The findings offer a roadmap for the development of new therapies to target the condition.
Schizophrenia is an often devastating disorder causing huge human suffering. Genetic studies have linked hundreds of genes to schizophrenia, each contributing a small part to the risk of developing the disease. The great abundance of identified genes have made it difficult to design experiments. Scientists have been struggling to understand what is linking the genes together and whether these genes affect the entire brain diffusely or certain components more.
By combining new maps of all the genes used in different cell types in the brain with detailed lists of the genes associated with schizophrenia, scientists in the current study could identify the types of cells that underlie the disorder. The genetics point towards certain cell types being much more implicated than others. One finding was that there appear to be a few major cell types contributing to the disorder, each of which originates in distinct areas of the brain.
“This marks a transition in how we can use large genetic studies to understand the biology of disease. With the results from this study, we are giving the scientific community a chance to focus their efforts where it will give maximum effect”, says Jens Hjerling-Leffler, research group leader at the Department of Medical Biochemistry and Biophysics at Karolinska Institutet, one of the main authors.
The findings offer a roadmap for the development of new therapies.
“One question now is whether these brain cell types are related to the clinical features of schizophrenia. For example, greater dysfunction in one cell type could make treatment response less likely. Dysfunction in a different cell type could increase the chances of long-term cognitive effects. This would have important implications for the development of new treatments, as separate drugs may be required for each cell type involved,” says co-main author Patrick Sullivan, Professor at the Department of Medical Epidemiology and Biostatistics at Karolinska Institutet and Yeargan Distinguished Professor in the Department of Genetics and Psychiatry at the University of North Carolina.
As a result of rapid progress in two separate fields of science; human genetics and single-cell transcriptomics, it only recently has become possible to study diseases in this way. In coming years the researchers suggest that the approach should lead to breakthroughs in the biological understanding of other complex disorders such as autism, major depression, and eating disorders.
“Understanding which cell types are affected in a disease is of critical importance for developing new medicines to improve their treatment. If we do not know what causes a disorder we cannot study how to treat it,” says Nathan Skene, Postdoc at the Department of Medical Biochemistry and Biophysics at Karolinska Institutet and UCL Institute of Neurology, UK, one of the lead authors.
Reference: “Genetic identification of brain cell types underlying schizophrenia” by Nathan G. Skene, Julien Bryois, Trygve E. Bakken, Gerome Breen, James J. Crowley, Héléna A. Gaspar, Paola Giusti-Rodriguez, Rebecca D. Hodge, Jeremy A. Miller, Ana B. Muñoz-Manchado, Michael C. O’Donovan, Michael J. Owen, Antonio F. Pardiñas, Jesper Ryge, James T. R. Walters, Sten Linnarsson, Ed S. Lein, Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Patrick F. Sullivan and Jens Hjerling-Leffler, 21 May 2018, Nature Genetics.
DOI: 10.1038/s41588-018-0129-5






Schizophrenia is a really big problem in the world