
Researchers in Frankfurt and Jena have now deciphered how the disturbed recycling chain of the endoplasmic reticulum can cause neurodegenerative diseases. Credit: Manja Schiefer for Jena University Hospital
Researchers have discovered the mechanisms that regulate the structure and function of the endoplasmic reticulum.
The endoplasmic reticulum, often abbreviated as ER, is a complex network of tubes, sacs, and membrane-bound compartments that pervade the cells of humans, animals, plants, and fungi. It serves as the manufacturing hub for proteins, overseeing their production, ensuring they fold into the appropriate three-dimensional structure, and modifying them as needed. Additionally, the ER is integral to the production of lipids and hormones, and is responsible for maintaining the cell’s calcium balance.
In addition, the ER serves as the foundation for the cell’s transport system, facilitating the movement of materials within the cellular environment. It also plays a key role in quality control by directing misfolded proteins toward the cell’s internal waste disposal system. Furthermore, it neutralizes harmful toxins that find their way into the cell, thus safeguarding the cell’s functionality and health.
In view of its multiple tasks, the ER is constantly being remodeled. A process called ER-phagy (roughly “self-digestion of the ER”) is responsible for ER degradation. Involved is a group of signal-receiving proteins – receptors – that are responsible for the membrane curvatures of the ER and thus for its multiple forms in the cell.
In ER-phagy, the receptors accumulate at specific sites on the ER and increase membrane curvature to such an extent that, as a consequence, part of the ER is strangulated and broken down into its component parts by cellular recycling structures (autophagosomes).
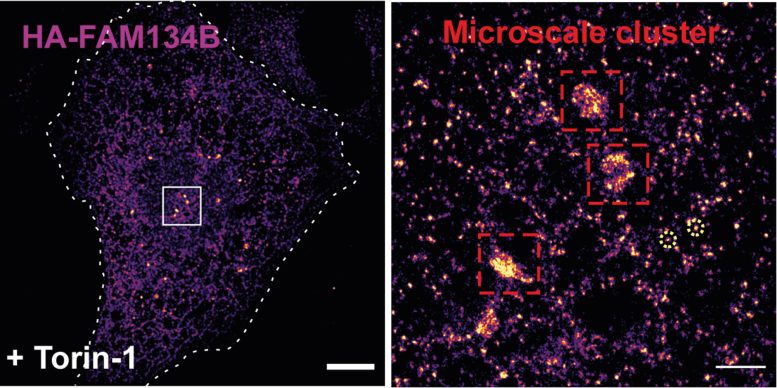
A super-high resolution microscopy technique reveals how FAM134B proteins assemble into clusters after stimulation of ER-phagy in the endoplasmic reticulum. Credit: Gonzáles et al., Nature (2023)
In cell culture experiments, biochemical and molecular biological studies, and computer simulations, the scientific team led by Professor Ivan Đikić of Goethe University Frankfurt first tested the membrane curvature receptor FAM134B and demonstrated that ubiquitin promotes and stabilizes the formation of clusters of FAM134B protein in the ER membrane.
Thus, ubiquitin drives ER-phagy. Đikić explains: “Ubiquitin causes the FAM134B clusters to become more stable and the ER to bulge out more at these sites. The stronger membrane curvature then leads to further stabilization of the clusters and, moreover, attracts additional membrane curvature proteins. So the effect of ubiquitin is self-reinforcing.” The researchers were also able to detect cluster formation using super-high-resolution microscopy.
Đikić continues: “To fulfill this function, ubiquitin changes the shape of part of the FAM134B protein. This is another facet of ubiquitin that performs an almost unbelievable array of tasks to keep all different cell functions working.”
The importance of ER-phagy is demonstrated by diseases resulting from a defective FAM134B protein. A team led by Professor Christian Hübner from Jena University Hospital previously identified mutations in the FAM134B gene causing a very rare hereditary sensory and autonomic neuropathy (HSAN), in which sensory nerves die. As a result, patients are unable to perceive pain and temperature correctly, which can lead to incorrect stresses or injuries going unnoticed and developing into chronic wounds. In a long-standing collaboration between Jena University Hospital and Goethe University Frankfurt FAM134B was identified as the first receptor for ER-phagy.
Mutations in another membrane curvature protein called ARL6IP1 cause a similar neurodegenerative disorder which combines sensory defects with muscle hardening (spasticity) in the legs. The scientific team led by Christian Hübner and Ivan Đikić has now identified that ARL6IP1 belongs to the ER-phagy machinery as well and is also ubiquitinated during ER-phagy.
Christian Hübner explains: “In mice that do not possess the ARL6IP1 protein, we can see that the ER virtually expands and degenerates as the cells age. This leads to an accumulation of misfolded proteins or protein clumps, which are no longer disposed of in the cell. As a result, nerve cells in particular, which do not renew as quickly as other body cells, die, causing the clinical symptoms in affected patients and genetically modified mice. We hypothesize from our data that the two membrane curvature receptors FAM134B and ARL6IP1 form mixed clusters during ER-phagy and depend on each other to control normal size and function of ER. Additional work will be required to fully acknowledge the role of ER-phagy in neurons as well as in other cell types.”
Overall, however, the research teams have taken a decisive step toward understanding ER-phagy, Đikić is convinced: “We now understand better how cells control their functions and thus create something we call cellular homeostasis. In biology, this knowledge allows fascinating insights into the incredible achievements of our cells, and for medicine it is essential for understanding diseases, diagnosing them on time, and helping patients by developing new therapies.”
References: “Ubiquitination regulates ER-phagy and remodeling of endoplasmic reticulum” by Alexis González, Adriana Covarrubias-Pinto, Ramachandra M. Bhaskara, Marius Glogger, Santosh K. Kuncha, Audrey Xavier, Eric Seemann, Mohit Misra, Marina E. Hoffmann, Bastian Bräuning, Ashwin Balakrishnan, Britta Qualmann, Volker Dötsch, Brenda A. Schulman, Michael M. Kessels, Christian A. Hübner, Mike Heilemann, Gerhard Hummer and Ivan Dikić, 24 May 2023, Nature.
DOI: 10.1038/s41586-023-06089-2
“Heteromeric clusters of ubiquitinated ER-shaping proteins drive ER-phagy” by Hector Foronda, Yangxue Fu, Adriana Covarrubias-Pinto, Hartmut T. Bocker, Alexis González, Eric Seemann, Patricia Franzka, Andrea Bock, Ramachandra M. Bhaskara, Lutz Liebmann, Marina E. Hoffmann, Istvan Katona, Nicole Koch, Joachim Weis, Ingo Kurth, Joseph G. Gleeson, Fulvio Reggiori, Gerhard Hummer, Michael M. Kessels, Britta Qualmann, Muriel Mari, Ivan Dikić and Christian A. Hübner, 24 May 2023, Nature.
DOI: 10.1038/s41586-023-06090-9



Be the first to comment on "When the Cell Digests Itself – Unraveling the Origins of Neurodegenerative Diseases"