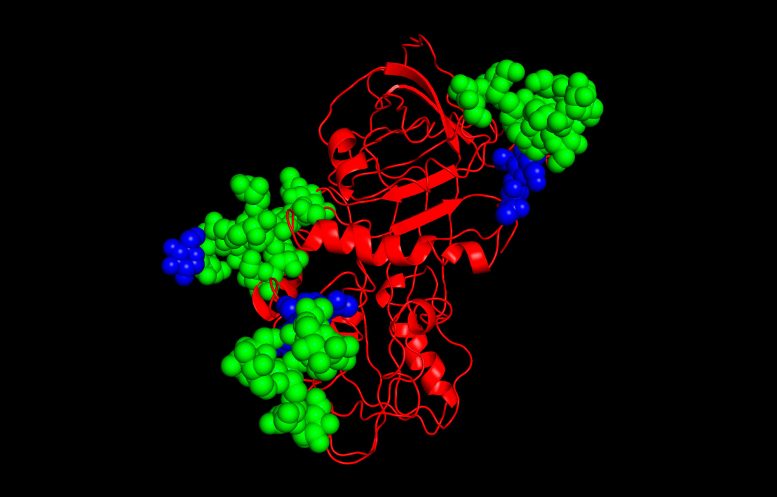
A groundbreaking study at UMass Amherst has decoded how sugars attached to proteins guide their correct folding, shedding light on potential treatments for diseases caused by protein misfolding. This protein (red) has been glycosolated with glycans (blue and green). Credit: UMass Amherst
Team’s approach reveals crucial role played by a specific enzyme in the folding process.
While we often think of diseases as caused by foreign bodies—bacteria or viruses—there are hundreds of diseases affecting humans that result from errors in cellular production of its proteins. A team of researchers led by the University of Massachusetts Amherst recently leveraged the power of cutting-edge technology, including an innovative technique called glycoproteomics, to unlock the carbohydrate-based code that governs how certain classes of proteins form themselves into the complex shapes necessary to keep us healthy.
Unraveling the Mysteries of Serpins
The research, published in the journal Molecular Cell, explores members of a family of proteins called serpins, which are implicated in a number of diseases. The research is the first to investigate how the location and composition of carbohydrates attached to the serpins ensure that they fold correctly. Serious diseases—ranging from emphysema and cystic fibrosis to Alzheimer’s disease—can result when the cellular oversight of protein folding goes awry. Identifying the glyco-code responsible for high-fidelity folding and quality control could be a promising way for drug therapies to target many diseases.
DNA and Beyond: Understanding Protein Folding
Scientists once thought that the single code governing life was DNA, and that everything was governed by how DNA’s four building blocks—A, C, G and T—combined and recombined. But in recent decades, it has become clear that there are other codes at work, and especially in building the intricately folded, secreted proteins that are created in the human cell’s protein factory, the endoplasmic reticulum (ER), a membrane-enclosed compartment where protein folding begins.
Approximately 7,000 different proteins—one third of all the proteins in the human body—mature in the ER. The secreted proteins—collectively known as the “secretome”—are responsible for everything from our body’s enzymes to its immune and digestive systems and must be formed correctly for the human body to function normally.
The Role of Chaperones in Protein Folding
Special molecules called “chaperones,” help fold the protein into its final shape. They also help to identify proteins that haven’t folded quite correctly, lending them additional help in refolding, or, if they’re hopelessly misfolded, targeting them for destruction before they cause damage. However, the chaperone system itself, which comprises a part of the cell’s quality control department, sometimes fails, and when it does, the results can be catastrophic for our health.
Pioneering Discoveries in Glycoproteomics
The discovery of the carbohydrate-based chaperone system in the ER was due to the pioneering work that Daniel Hebert, professor of biochemistry and molecular biology at UMass Amherst and one of the paper’s senior authors, initiated as a postdoctoral fellow in the 1990s. “The tools we have now, including glycoproteomics and mass spectrometry at UMass Amherst’s Institute for Applied Life Sciences, are allowing us to answer questions that have remained open for over 25 years,” says Hebert. “The lead author of this new paper, Kevin Guay, is doing things I could only dream of when I first started.”
Among the most pressing of these unanswered questions is: how do chaperones know when 7,000 different origami-like proteins are correctly folded?
Innovations in Understanding Protein Quality Control
We know now that the answer involves an “ER gatekeeper” enzyme known as UGGT, and a host of carbohydrate tags, called N-glycans, which are linked to specific sites in the protein’s amino acid sequence.
Guay, who is completing his Ph.D. in the molecular cellular biology program at UMass Amherst, focused on two specific mammalian proteins, known as alpha-1 antitrypsin and antithrombin. Using CRISPR-edited cells, he and his co-authors modified the ER chaperone network to determine how the presence and location of N-glycans affected protein folding. They watched as the disease variants were recognized by the ER gatekeeper UGGT and, in order to peer more closely, developed a number of innovative glycoproteomics techniques using mass spectrometry to understand what happens to the glycans that stud the surface of the proteins.
What they discovered is that the enzyme UGGT “tags” misfolded proteins with sugars placed in specific positions. It’s a sort of code that the chaperones can then read to determine exactly where the folding process went wrong and how to fix it.
Implications and Future Directions
“This is the first time that we’ve been able to see where UGGT puts sugars on proteins made in human cells for quality control,” says Guay. “We now have a platform for extending our understanding of how sugar tags can send proteins for further quality control steps and our work suggests that UGGT is a promising avenue for targeted drug therapy research.”
“What’s so exciting about this research,” says Lila Gierasch, distinguished professor of biochemistry and molecular biology at UMass Amherst and one of the paper’s co-authors, “is the discovery that glycans act as a code for protein folding in the ER. The discovery of the role that UGGT plays opens the door to future advancement in understanding and eventually treating the hundreds of diseases that result from misfolded proteins.”
Reference: “ER chaperones use a protein folding and quality control glyco-code” by Kevin P. Guay, Haiping Ke, Nathan P. Canniff, Gracie T. George, Stephen J. Eyles, Malaiyalam Mariappan, Joseph N. Contessa, Anne Gershenson, Lila M. Gierasch and Daniel N. Hebert, 4 December 2023, Molecular Cell.
DOI: 10.1016/j.molcel.2023.11.006





Well above my education and skill levels, the research being described is absolutely amazing. However, equally amazing perhaps, as a long-term lay victim of some common chronic illnesses merely trying to survive and thrive in the midst of so much mainstream medical ignorance, incompetence and erring, I apparently discovered some of the causes of that protein misfolding by late 1981.
Mysteriously, seriously ill with chronic fatigue, generalized aches, pains and muscle weakness, serious mood swings, asymptomatic gout and a low oral temperature in early 1981, by late 1981 I learned I was/am mildly allergic to many common foods (e.g., cow’s milk; inherited?) and food additives (e.g., MSG; FDA approval, 1980) through shortly thereafter medically abandoned “cytotoxic blood testing for food allergies.”
While understanding the protein misfolding/destruction process may eventually lead to valuable treatments, learning of and partially avoiding one’s allergies and some illegally FDA approved food additives can provide immediate benefit. Meanwhile, I posit that learning how high serum levels of uric acid affects protein folding may ultimately prove to be invaluable.