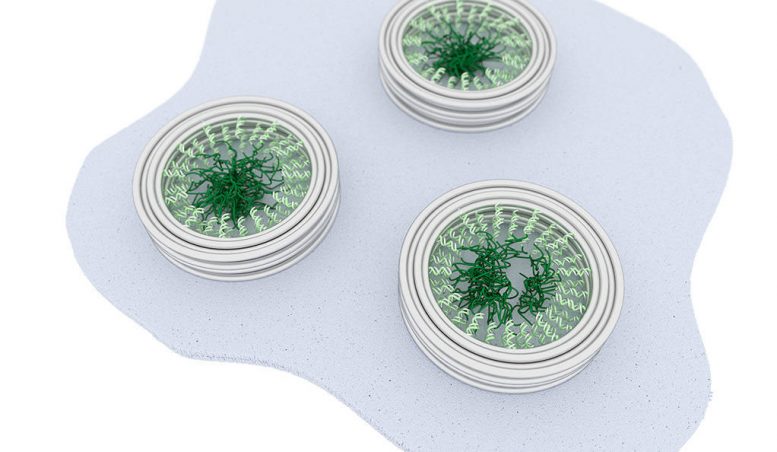
Yale researchers replicated a nuclear membrane channel and observed “molecular bouncers” regulating access to it, shedding light on how molecules move in cells.
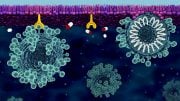
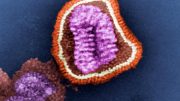
DNA is packaged tightly within the cell’s nuclear membranes, which contain channels that regulate the transit of macromolecules governing all of life’s functions. Yale University researchers have built a nanoscale replica of this channel and have visualized the interaction of proteins that act as “molecular bouncers,” controlling access to the channel’s 40-nanometer entrance.
“In the past, we have tried to break things apart to study them, but if you really want to understand how these channels work, you should be able to build them,” said Patrick Lusk, associate professor of cell biology at Yale School of Medicine.
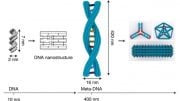
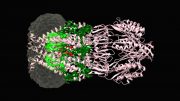
In collaboration with Chenxiang Lin, an assistant professor of cell biology and a member of the Nanobiology Institute at Yale’s West Campus, the researchers recreated key aspects of the nuclear membrane transport channels by meticulously arranging the native cellular proteins on a nano-cylinder made, constructed using a technique called “DNA-origami.” They managed to capture images of the protein interactions, seen within the cylinder in the accompanying video.
Reference: “A Programmable DNA Origami Platform for Organizing Intrinsically Disordered Nucleoporins within Nanopore Confinement” by Patrick D. Ellis Fisher, Qi Shen, Bernice Akpinar, Luke K. Davis, Kenny Kwok Hin Chung, David Baddeley, Anđela Šarić, Thomas J. Meli†, Bart W. Hoogenboom, Chenxiang Lin and C. Patrick Lusk, 19 January 2018, ACS Nano.
DOI: 10.1021/acsnano.7b08044



Very informative post related to bioinformatics. It is great help for my new project .By reading this article, I learn some important things that I need to improve. I continuously check this site for regular updates in field. Thanks for putting top notch content in article. I would like to be here again to find another masterpiece article.
I am so grateful to enter this freecell for the time i spend with time, it was a great experience and did help to develop my patience. the more than exciting levels are available in this game.