
A groundbreaking study maps the genetic relationships of over 9,500 flowering plant species, creating an advanced tree of life that enhances our understanding of their evolutionary history and potential uses in various scientific fields. Credit: SciTechDaily.com
The largest-ever tree of life for flowering plants has been constructed by sequencing the DNA of more than 9,500 species, charting the evolutionary and genetic connections among these plants.
A recent study published in the journal Nature, authored by an international team of 279 researchers, including three scientists from the New York Botanical Garden (NYBG), offers the latest insights into the evolutionary and genetic relationships among flowering plants. These plants account for approximately 90 percent of all known plant species.
Using 1.8 billion letters of genetic code from over 9,500 species covering almost 8,000 plant genera (groups of closely related species), the research team was able to create the most detailed tree of life—a graphic depiction of species relationships similar to a genealogical family tree—to date for this group of plants, shedding new light on the evolutionary history of flowering plants and their rise to ecological dominance on Earth. The study’s authors believe the data will aid future attempts to identify new species, refine plant classification, uncover new medicinal compounds, and conserve plants in the face of the dual biodiversity and climate crises.
Contributing to this major milestone in plant science were Fabián Michelangeli, Ph.D., Abess Curator of Tropical Botany and Director of NYBG’s Institute of Systematic Botany; Gregory M. Plunkett, Ph.D., Director and Curator of NYBG’s Cullman Program for Molecular Systematics; and John D. Mitchell, NYBG Affiliated Scientist.
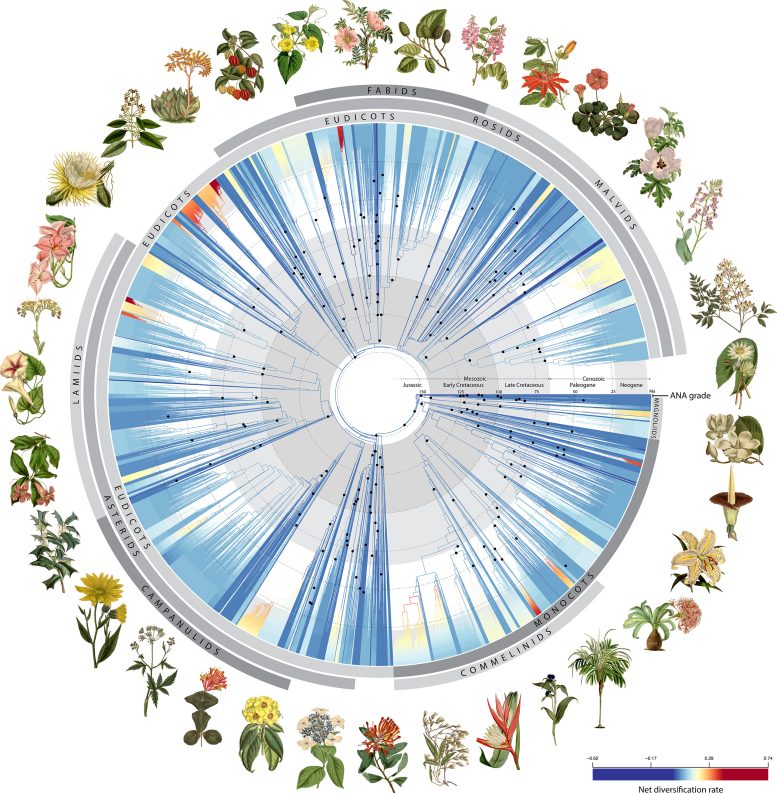
An international team of researchers, including three New York Botanical Garden (NYBG) scientists, used genetic code from more than 9,500 flowering plant species to create the most detailed evolutionary tree of life for this group of plants to date. Credit: RBG Kew
“While the main goals of this large-scale project were to understand the relationships of all flowering plant genera, it also sheds light on the timing of major events in the evolution of complex flower forms and life histories,” Dr. Michelangeli said. “Large analyses such as this can provide context for conservation strategies, sustainable agriculture, and many other applications that need basic biodiversity knowledge. Understanding how organisms are related is the building block of all biodiversity science and applications.”
The research team—led by the Royal Botanic Gardens, Kew, and involving 138 organizations internationally—used 15 times more data than any comparable studies of the flowering plant tree of life. Among the species included in the study, the DNA of more than 800 had never been sequenced before. The sheer amount of data unlocked by this research, which would take a single computer 18 years to process, is a huge stride towards building a tree of life for all 330,000 known species of flowering plants.
Drs. Michelangeli and Plunkett and Mr. Mitchell provided expertise on the plant families they study as well as expertly identified samples for a variety of plant groups, with a large proportion coming from the Melastomataceae family of tropical plants, which is Dr. Michelangeli’s specialty, and the Apiaceae (parsley or carrot) and Araliaceae (ginseng) families, which Dr. Plunkett studies.
Unlocking Historic Herbarium Specimens for Cutting-Edge Research
The flowering plant tree of life, much like a family tree, enables scientists to understand how different species are related to each other. The tree of life is uncovered by comparing DNA sequences between different species to identify changes (mutations) that accumulate over time like a molecular fossil record. Science’s understanding of the tree of life is improving rapidly in tandem with advances in DNA-sequencing technology. For this study, new genomic techniques were developed to magnetically capture hundreds of genes and hundreds of thousands of letters of genetic code from every sample, orders of magnitude more than earlier methods.
A key advantage of the team’s approach is that it enables a wide diversity of plant material, old and new, to be sequenced, even when the DNA is badly damaged. The vast treasure troves of dried, preserved plants in the world’s herbarium collections, which comprise nearly 400 million specimens, can now be studied genetically. Using such specimens, the team successfully sequenced a sandwort (Arenaria globiflora) collected nearly 200 years ago in Nepal and, despite the poor quality of its DNA, were able to place it on the tree of life. The team even analyzed extinct plants, such has the Guadalupe Island olive (Hesperelaea palmeri), which has not been seen alive since 1875. In fact, 511 of the species sequenced are already at risk of extinction, according to the Red List, the authoritative compilation of the world’s threatened plant, fungal, and animal species maintained by the International Union for Conservation of Nature.
Across all 9,506 species sequenced, over 3,400 came from material sourced from 163 herbaria in 48 countries. Additional material from plant collections around the world such as DNA banks, seeds, and living collections have been vital for filling key knowledge gaps to shed new light on the history of flowering plant evolution. The team also benefited from publicly available data for over 1,900 species, highlighting the value of the open-science approach to future genomic research.
Illuminating Darwin’s “Abominable Mystery”
Flowering plants account for about 90 percent of all known plant life on land and are found virtually everywhere on the planet—from the steamiest tropics to the rocky outcrops of the Antarctic Peninsula. And yet our understanding of how these plants came to dominate the scene soon after their origin has baffled scientists for generations, including Charles Darwin. Flowering plants originated over 140 million years ago after which they rapidly overtook other vascular plants, including their closest living relatives—the gymnosperms, non-flowering plants that have naked seeds such as cycads, conifers, and ginkgo.
Darwin was mystified by the seemingly sudden appearance of such diversity in the fossil record. In an 1879 letter to Joseph Dalton Hooker, his close confidant and Director of the Royal Botanic Gardens, Kew, he wrote, “The rapid development as far as we can judge of all the higher plants within recent geological times is an abominable mystery.”
Using 200 fossils, the researchers scaled their tree of life to time, revealing how flowering plants evolved across geological time. They found that early flowering plants exploded in diversity, giving rise to over 80 percent of the major lineages that exist today shortly after their origin. However, this trend then declined to a steadier rate for the next 100 million years until another surge in diversification about 40 million years ago, coinciding with a global decline in temperatures. These new insights would have fascinated Darwin and will surely help today’s scientists grappling with the challenges of understanding how and why species diversify.
Assembling a tree of life this extensive would have been impossible without the collaboration of scientists across the globe. In total, 279 authors were involved in the research, representing many different nationalities from 138 organizations in 27 countries. International collaborators shared their unique botanical expertise as well as many invaluable plant samples from around the world that could not be obtained without their help. The comprehensive nature of the tree is in no small part a result of this wide-ranging partnership.
“Efforts like this show how the international scientific community can come together to collaborate and produce something that no one research group or institution can do alone,” Dr. Michelangeli said.
Putting the Flowering Plant Tree of Life to Good Use
The flowering plant tree of life has enormous potential in biodiversity research. This is because, just as one can predict the properties of an element based on its position in the periodic table, the location of a species in the tree of life allows scientists to predict its properties. The new data will thus be invaluable for enhancing many areas of science and beyond.
To enable this, the tree and all of the data that underpin it have been made openly and freely accessible to both the public and scientific community, including through the Kew Tree of Life Explorer. The study’s authors believe such open access is key to democratizing access to scientific data across the globe.
Open access will also help scientists to make the best use of the data such as combining it with artificial intelligence to predict which plant species may include molecules with medicinal potential. Similarly, the tree of life can be used to better understand and predict how pests and diseases might affect the world’s plants in the future. Ultimately, the authors note, the applications of the data will be driven by the ingenuity of scientists.
Reference: “Phylogenomics and the rise of the angiosperms” by Alexandre R. Zuntini, Tom Carruthers, Olivier Maurin, Paul C. Bailey, Kevin Leempoel, Grace E. Brewer, Niroshini Epitawalage, Elaine Françoso, Berta Gallego-Paramo, Catherine McGinnie, Raquel Negrão, Shyamali R. Roy, Lalita Simpson, Eduardo Toledo Romero, Vanessa M. A. Barber, Laura Botigué, James J. Clarkson, Robyn S. Cowan, Steven Dodsworth, Matthew G. Johnson, Jan T. Kim, Lisa Pokorny, Norman J. Wickett, Guilherme M. Antar, Lucinda DeBolt, Karime Gutierrez, Kasper P. Hendriks, Alina Hoewener, Ai-Qun Hu, Elizabeth M. Joyce, Izai A. B. S. Kikuchi, Isabel Larridon, Drew A. Larson, Elton John de Lírio, Jing-Xia Liu, Panagiota Malakasi, Natalia A. S. Przelomska, Toral Shah, Juan Viruel, Theodore R. Allnutt, Gabriel K. Ameka, Rose L. Andrew, Marc S. Appelhans, Montserrat Arista, María Jesús Ariza, Juan Arroyo, Watchara Arthan, Julien B. Bachelier, C. Donovan Bailey, Helen F. Barnes, Matthew D. Barrett, Russell L. Barrett, Randall J. Bayer, Michael J. Bayly, Ed Biffin, Nicky Biggs, Joanne L. Birch, Diego Bogarín, Renata Borosova, Alexander M. C. Bowles, Peter C. Boyce, Gemma L. C. Bramley, Marie Briggs, Linda Broadhurst, Gillian K. Brown, Jeremy J. Bruhl, Anne Bruneau, Sven Buerki, Edie Burns, Margaret Byrne, Stuart Cable, Ainsley Calladine, Martin W. Callmander, Ángela Cano, David J. Cantrill, Warren M. Cardinal-McTeague, Mónica M. Carlsen, Abigail J. A. Carruthers, Alejandra de Castro Mateo, Mark W. Chase, Lars W. Chatrou, Martin Cheek, Shilin Chen, Maarten J. M. Christenhusz, Pascal-Antoine Christin, Mark A. Clements, Skye C. Coffey, John G. Conran, Xavier Cornejo, Thomas L. P. Couvreur, Ian D. Cowie, Laszlo Csiba, Iain Darbyshire, Gerrit Davidse, Nina M. J. Davies, Aaron P. Davis, Kor-jent van Dijk, Stephen R. Downie, Marco F. Duretto, Melvin R. Duvall, Sara L. Edwards, Urs Eggli, Roy H. J. Erkens, Marcial Escudero, Manuel de la Estrella, Federico Fabriani, Michael F. Fay, Paola de L. Ferreira, Sarah Z. Ficinski, Rachael M. Fowler, Sue Frisby, Lin Fu, Tim Fulcher, Mercè Galbany-Casals, Elliot M. Gardner, Dmitry A. German, Augusto Giaretta, Marc Gibernau, Lynn J. Gillespie, Cynthia C. González, David J. Goyder, Sean W. Graham, Aurélie Grall, Laura Green, Bee F. Gunn, Diego G. Gutiérrez, Jan Hackel, Thomas Haevermans, Anna Haigh, Jocelyn C. Hall, Tony Hall, Melissa J. Harrison, Sebastian A. Hatt, Oriane Hidalgo, Trevor R. Hodkinson, Gareth D. Holmes, Helen C. F. Hopkins, Christopher J. Jackson, Shelley A. James, Richard W. Jobson, Gudrun Kadereit, Imalka M. Kahandawala, Kent Kainulainen, Masahiro Kato, Elizabeth A. Kellogg, Graham J. King, Beata Klejevskaja, Bente B. Klitgaard, Ronell R. Klopper, Sandra Knapp, Marcus A. Koch, James H. Leebens-Mack, Frederic Lens, Christine J. Leon, Étienne Léveillé-Bourret, Gwilym P. Lewis, De-Zhu Li, Lan Li, Sigrid Liede-Schumann, Tatyana Livshultz, David Lorence, Meng Lu, Patricia Lu-Irving, Jaquelini Luber, Eve J. Lucas, Manuel Luján, Mabel Lum, Terry D. Macfarlane, Carlos Magdalena, Vidal F. Mansano, Lizo E. Masters, Simon J. Mayo, Kristina McColl, Angela J. McDonnell, Andrew E. McDougall, Todd G. B. McLay, Hannah McPherson, Rosa I. Meneses, Vincent S. F. T. Merckx, Fabián A. Michelangeli, John D. Mitchell, Alexandre K. Monro, Michael J. Moore, Taryn L. Mueller, Klaus Mummenhoff, Jérôme Munzinger, Priscilla Muriel, Daniel J. Murphy, Katharina Nargar, Lars Nauheimer, Francis J. Nge, Reto Nyffeler, Andrés Orejuela, Edgardo M. Ortiz, Luis Palazzesi, Ariane Luna Peixoto, Susan K. Pell, Jaume Pellicer, Darin S. Penneys, Oscar A. Perez-Escobar, Claes Persson, Marc Pignal, Yohan Pillon, José R. Pirani, Gregory M. Plunkett, Robyn F. Powell, Ghillean T. Prance, Carmen Puglisi, Ming Qin, Richard K. Rabeler, Paul E. J. Rees, Matthew Renner, Eric H. Roalson, Michele Rodda, Zachary S. Rogers, Saba Rokni, Rolf Rutishauser, Miguel F. de Salas, Hanno Schaefer, Rowan J. Schley, Alexander Schmidt-Lebuhn, Alison Shapcott, Ihsan Al-Shehbaz, Kelly A. Shepherd, Mark P. Simmons, André O. Simões, Ana Rita G. Simões, Michelle Siros, Eric C. Smidt, James F. Smith, Neil Snow, Douglas E. Soltis, Pamela S. Soltis, Robert J. Soreng, Cynthia A. Sothers, Julian R. Starr, Peter F. Stevens, Shannon C. K. Straub, Lena Struwe, Jennifer M. Taylor, Ian R. H. Telford, Andrew H. Thornhill, Ifeanna Tooth, Anna Trias-Blasi, Frank Udovicic, Timothy M. A. Utteridge, Jose C. Del Valle, G. Anthony Verboom, Helen P. Vonow, Maria S. Vorontsova, Jurriaan M. de Vos, Noor Al-Wattar, Michelle Waycott, Cassiano A. D. Welker, Adam J. White, Jan J. Wieringa, Luis T. Williamson, Trevor C. Wilson, Sin Yeng Wong, Lisa A. Woods, Roseina Woods, Stuart Worboys, Martin Xanthos, Ya Yang, Yu-Xiao Zhang, Meng-Yuan Zhou, Sue Zmarzty, Fernando O. Zuloaga, Alexandre Antonelli, Sidonie Bellot, Darren M. Crayn, Olwen M. Grace, Paul J. Kersey, Ilia J. Leitch, Hervé Sauquet, Stephen A. Smith, Wolf L. Eiserhardt, Félix Forest and William J. Baker, 24 April 2024, Nature.
DOI: 10.1038/s41586-024-07324-0







Be the first to comment on "Groundbreaking New Tree of Life Sheds Light on Darwin’s “Abominable Mystery”"