The X chromosome is the first human chromosome to be completely sequenced from end to end, with no gaps in the sequence and an unprecedented level of accuracy.
The first end-to-end (‘telomere-to-telomere’) completely gapless DNA sequence of a human chromosome is a major milestone for genomics research.
Although the current human reference genome is the most accurate and complete vertebrate genome ever produced, there are still gaps in the DNA sequence, even after two decades of improvements. Now, for the first time, scientists have determined the complete sequence of a human chromosome from one end to the other (‘telomere to telomere’) with no gaps and an unprecedented level of accuracy.
The publication of the telomere-to-telomere assembly of a complete human X chromosome on July 14, 2020, in Nature is a landmark achievement for genomics researchers. Lead author Karen Miga, a research scientist at the UC Santa Cruz Genomics Institute, said the project was made possible by new sequencing technologies that enable “ultra-long reads,” such as the nanopore sequencing technology pioneered at UC Santa Cruz.
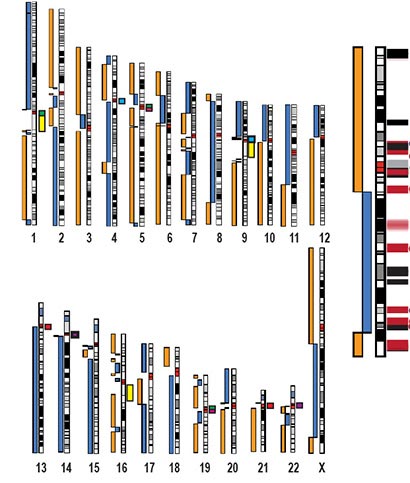
Chromosome ideograms showing the whole-genome assembly of the human CHM13 genome, exceeding all prior human genome assemblies in terms of continuity, completeness, and accuracy. The X chromosome is enlarged on the right. Gapless contigs are illustrated as blue and orange bars next to the chromosome ideograms. Credit: Miga et al., Nature 2020
Repetitive DNA sequences are common throughout the genome and have always posed a challenge for sequencing because most technologies produce relatively short “reads” of the sequence, which then have to be pieced together like a jigsaw puzzle to assemble the genome. Repetitive sequences yield lots of short reads that look almost identical, like a large expanse of blue sky in a puzzle, with no clues as to how the pieces fit together or how many repeats there are.
“These repeat-rich sequences were once deemed intractable, but now we’ve made leaps and bounds in sequencing technology,” Miga said. “With nanopore sequencing, we get ultra-long reads of hundreds of thousands of base pairs that can span an entire repeat region, so that bypasses some of the challenges.”
Filling in the remaining gaps in the human genome sequence opens up new regions of the genome where researchers can search for associations between sequence variations and disease and for other clues to important questions about human biology and evolution.
“We’re starting to find that some of these regions where there were gaps in the reference sequence are actually among the richest for variation in human populations, so we’ve been missing a lot of information that could be important to understanding human biology and disease,” Miga said.
Telomere to telomere
Miga and Adam Phillippy at the National Human Genome Research Institute (NHGRI), both corresponding authors of the new paper, co-founded the Telomere-to-Telomere (T2T) consortium to pursue a complete genome assembly after working together on a 2018 paper that demonstrated the potential of nanopore technology to produce a complete human genome sequence. That effort used the Oxford Nanopore Technologies MinION sequencer, which sequences DNA by detecting the change in current flow as single molecules of DNA pass through a tiny hole (a “nanopore”) in a membrane.

Research scientist Karen Miga at the UCSC Genomics Institute led the telomere-to-telomere assembly of a complete human X chromosome. Credit: UC Santa Cruz
The new project built on that effort, combining nanopore sequencing with other sequencing technologies from PacBio and Illumina, and optical maps from BioNano Genomics. Using these technologies, the team produced a whole-genome assembly that exceeds all prior human genome assemblies in terms of continuity, completeness, and accuracy, even surpassing the current human reference genome by some metrics.
Nevertheless, there were still multiple breaks in the sequence, Miga said. To finish the X chromosome, the team had to manually resolve several gaps in the sequence. Two segmental duplications were resolved with ultra-long nanopore reads that completely spanned the repeats and were uniquely anchored on either side. The remaining break was at the centromere, a notoriously difficult region of repetitive DNA found in every chromosome.
In the X chromosome, the centromere encompasses a region of highly repetitive DNA spanning 3.1 million base pairs (the bases A, C, T, and G form pairs in the DNA double helix and encode genetic information in their sequence). The team was able to identify variants within the repeat sequence to serve as markers, which they used to align the long reads and connect them together to span the entire centromere.
“For me, the idea that we can put together a 3-megabase-size tandem repeat is just mind-blowing. We can now reach these repeat regions covering millions of bases that were previously thought intractable,” Miga said.
Polishing strategy
The next step was a polishing strategy using data from multiple sequencing technologies to ensure the accuracy of every base in the sequence.
“We used an iterative process over three different sequencing platforms to polish the sequence and reach a high level of accuracy,” Miga explained. “The unique markers provide an anchoring system for the ultra-long reads, and once you anchor the reads, you can use multiple data sets to call each base.”
Nanopore sequencing, in addition to providing ultra-long reads, can also detect bases that have been modified by methylation, an “epigenetic” change that does not alter the sequence but has important effects on DNA structure and gene expression. By mapping patterns of methylation on the X chromosome, the team was able to confirm previous observations and reveal some intriguing trends in methylation patterns within the centromere.
The new human genome sequence, derived from a human cell line called CHM13, closes many gaps in the current reference genome, known as Genome Reference Consortium build 38 (GRCh38).
The T2T consortium is continuing to work toward completion of all of the CHM13 chromosomes. “It’s an open consortium, so in many respects this is a community-driven project, with a lot of people dedicating time and resources to it,” Miga said.
Read First End-to-End DNA Sequence of a Human Chromosome for more on this breakthrough.
Reference: “Telomere-to-telomere assembly of a complete human X chromosome” by Karen H. Miga, Sergey Koren, Arang Rhie, Mitchell R. Vollger, Ariel Gershman, Andrey Bzikadze, Shelise Brooks, Edmund Howe, David Porubsky, Glennis A. Logsdon, Valerie A. Schneider, Tamara Potapova, Jonathan Wood, William Chow, Joel Armstrong, Jeanne Fredrickson, Evgenia Pak, Kristof Tigyi, Milinn Kremitzki, Christopher Markovic, Valerie Maduro, Amalia Dutra, Gerard G. Bouffard, Alexander M. Chang, Nancy F. Hansen, Amy B. Wilfert, Françoise Thibaud-Nissen, Anthony D. Schmitt, Jon-Matthew Belton, Siddarth Selvaraj, Megan Y. Dennis, Daniela C. Soto, Ruta Sahasrabudhe, Gulhan Kaya, Josh Quick, Nicholas J. Loman, Nadine Holmes, Matthew Loose, Urvashi Surti, Rosa ana Risques, Tina A. Graves Lindsay, Robert Fulton, Ira Hall, Benedict Paten, Kerstin Howe, Winston Timp, Alice Young, James C. Mullikin, Pavel A. Pevzner, Jennifer L. Gerton, Beth A. Sullivan, Evan E. Eichler and Adam M. Phillippy, 14 July 2020, Nature.
DOI: 10.1038/s41586-020-2547-7
In addition to Miga and Phillippy, the authors of the paper include co-first author Sergey Koren at the National Human Genome Research Institute and scientists at nearly two dozen institutions in the U.S. and U.K., including the University of Washington, Johns Hopkins University, UC San Diego, and the Wellcome Sanger Institute. This work was supported by the U.S. National Institutes of Health.








Be the first to comment on "Landmark Achievement for Genomics Researchers: First Complete Assembly of Human X Chromosome"