
A Rhagoletis pomonella fly explores the native host fruits of hawthorn trees. Credit: Hannes Schuler
By applying the latest tools of genome sequencing and analysis to preserved evidence from experiments carried out at Notre Dame in the 1990s, a new evolution study finds a massive genome shift in one generation.
A team of biologists from Rice University, the University of Notre Dame, and three other schools has discovered that an agricultural pest that began plaguing U.S. apple growers in the 1850s likely did so after undergoing extensive and genome-wide changes in a single generation.
This new result, which appears online this week in the journal Ecology Letters, came from applying the latest tools of genome sequencing and analysis to preserve evidence from experiments carried out at Notre Dame in the 1990s. The research focuses on the fruit fly Rhagoletis pomonella, aka the “apple maggot,” a North American native that lays its eggs inside the fruit of the hawthorn tree. In the 1850s, a splinter group of Rhagoletis began laying eggs in apples in upstate New York, a move that required the flies to adapt the timing of their annual egg-laying cycle to match the fruiting time of their new hosts.
“Today, there are two forms of Rhagoletis pomonella in the U.S., the ancestral form that times its life cycle to the hawthorn tree and a derived form that is timed to apple trees, which fruit about 3-4 weeks earlier,” said study lead author Scott Egan, an evolutionary biologist in the Department of BioSciences at Rice. “These two forms have evolved very distinct differences and are on the path to evolving into two new species, and because they have a single generation per year, we know that all of the differences between the two has happened in no more than 170 generations.”
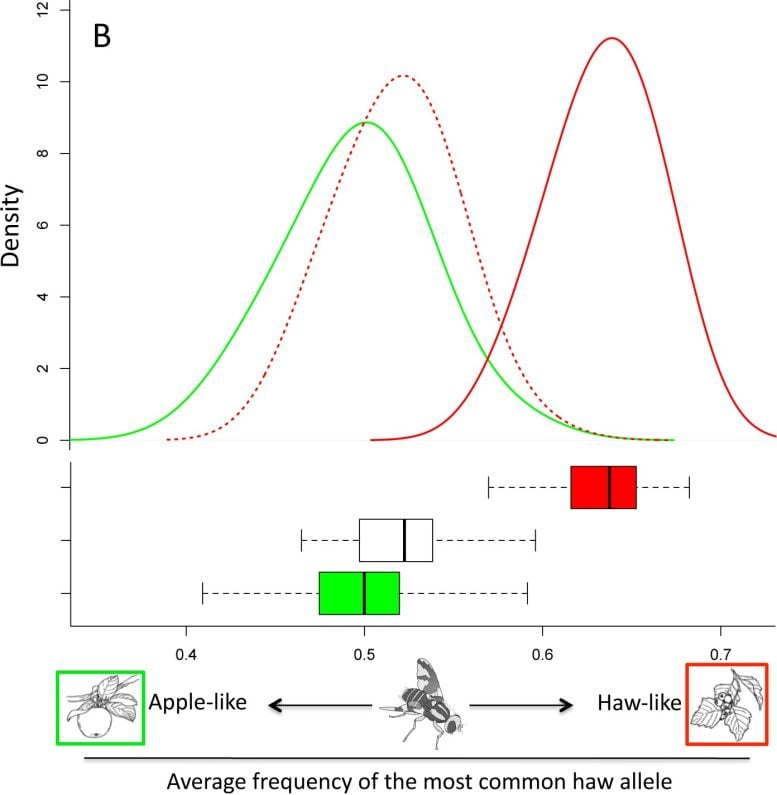
This graph depicts the genome-wide genetic differences that have evolved between a native host race of Rhagoletis pomonella that lays its eggs on hawthorn trees (red) and a new divergent host race (green) that began reproducing on apples trees in the 1850s. In laboratory experiments, researchers found that raising native haw flies on the apple trees’ fruiting cycle for just one generation led to a genome-wide shift (dashed red line) that accounted for up to 70 percent of the genetic differences between the two races. Credit: S. Egan/Rice University
The two populations are also known to interbreed. In a given year, about 5 percent move between populations. For Egan and other biologists who specialize in studying the process of speciation, continued “gene flow” between the divergent populations raises significant questions about whether and how R. pomonella may eventually evolve into two species.
“According to theory, this amount of gene flow should be enough to counteract the processes of divergent selection that are driving the populations apart,” Egan said. “However, if natural selection is strong, and the impacts of selection are genome-wide, the two might diverge even in the face of this much continued gene flow.”
To get a sense of how quickly the two populations diverged genetically, Egan and colleagues from Notre Dame, the University of Sheffield in Great Britain, the University of Florida, and Kansas State University conducted the most extensive genetic analysis of R. pomonella yet undertaken.
Borrowing a page from criminal forensics, the team applied the latest tools of genomic science to experimental samples that had been preserved in a freezer at the lab of Notre Dame biologist Jeffrey Feder since the mid-1990s. At the time of the initial experiment, genetic technology only allowed Feder and colleagues to study a handful of changes across the genome.
“We collected flies from native hawthorn trees, divided the flies into two groups and raised the flies under haw- and apple-like conditions in the lab,” Feder said. By raising the ancestral population under both conditions, Feder and colleagues experimentally replicated the host shift observed in New York in the 1850s.
In 2012, Egan and colleagues sequenced a portion of the genome for individuals from the earlier lab experiment and measured the differences between individuals that had been raised under haw- versus apple-like conditions. The team compiled a database of more than 5 billion nucleotide base pairs to examine the genetic differences between all the populations. Of particular interest were the sites across the genome where the DNA sequences differed from one population to the other at a single site. This type of genetic variation within a species is called a polymorphism, and “single nucleotide polymorphisms,” or SNPs, are the smallest unit of polymorphic variation.
“For the ancestral flies that had been raised for one generation on the apple cycle, we documented changes in more than 32,000 SNPs,” Egan said. “Overall, we found that the genetic changes undergone by this first generation accounted for up to 70 percent of all the genetic changes that had occurred between the two populations since the 1850s.”
Kansas State University’s Greg Ragland, co-lead author of the study, said, “The changes observed in both experimental and natural populations of R. pomonella underscore the importance of ecological selection at early stages of divergence and call for further integration of studies of speciation and genome divergence.”
For example, when the team compared the genetic changes from the “selection experiment” with the genome-wide differences between apple and haw populations in nature, they found striking similarities. Specifically, the direction and magnitude of the 32,000 SNPs were similar to the observed differences between the ancestral and divergent populations in nature.
“By combining genomic data from experimental and natural populations, we were able to quantify the total genome-wide impact of selection,” Feder said.
He said the effects are widespread across the R. pomonella genome because, in addition to direct effects on genes that underlie important traits, there were also indirect effects in which changes in one gene prompted changes elsewhere through a process called “linkage disequilibrium.”
Egan said previous research on R. pomonella provided other clues as to why the genetic shift was so widespread. For example, the traits that were under selection in the move between host plants involved many genes. In addition, R. pomonella has high levels of within-population genetic variation upon which selection easily acts. And finally, R. pomonella is more susceptible to the impact of divergent natural selection because its genome contains several “inversion areas,” regions where DNA sequences are broken and realigned in the opposite order, which increases linkage disequilibrium and the total impact of selection.
Reference: “Experimental evidence of genome-wide impact of ecological selection during early stages of speciation-with-gene-flow” by Scott P. Egan, Gregory J. Ragland, Lauren Assour, Thomas H.Q. Powell, Glen R. Hood, Scott Emrich, Patrik Nosil and Jeffrey L. Feder, 15 June 2015, Ecology Letters.
DOI: 10.1111/ele.12460
The study was funded by Rice University’s Huxley Faculty Fellowship in Ecology and Evolutionary Biology and by the University of Notre Dame’s Advanced Diagnostics and Therapeutics and Environmental Change Initiatives. Study co-authors include Lauren Assour, Glen Hood and Scott Emrich, all of Notre Dame; Thomas Powell of the University of Florida; and Patrik Nosil of the University of Sheffield.










“We collected flies from native hawthorn trees, divided the flies into two groups and raised the flies under haw- and apple-like conditions in the lab,” Feder said. By raising the ancestral population under both conditions, Feder and colleagues experimentally replicated the host shift observed in New York in the 1850s.
So, the gene shift was SOLELY due the arrival of the apple ala Johnny Appleseed?
I dont think anyone disputes life is the ultiate predator needing only a catastrophic event to upset the established order and begin a new predation based on an adjusted geneome.