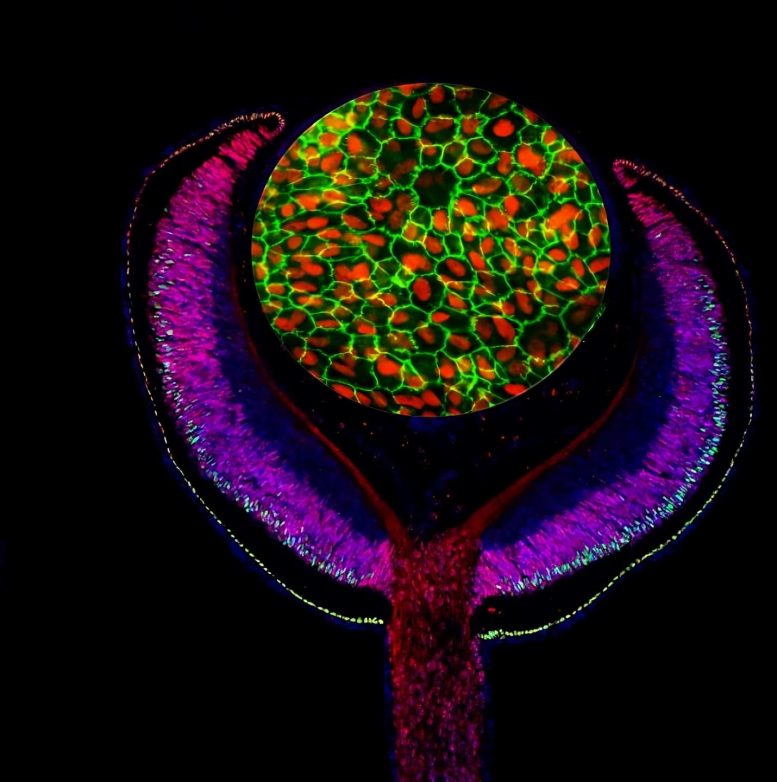
Composite of an embryonic mouse eye cup (E14.5) labeled with antibodies against the developmental transcription factors Lhx2 (red) and Otx2 (green), and cultured human retinal pigmented epithelium (RPE) labeled with antibodies against MITF (red) and ZO-1 (green). Credit: Images by Mazal Cohen-Gulkar, composite by Ruth Ashery-Padan (CC-BY 4.0)
Researchers have uncovered new insights into the causes of adult-onset macular degeneration.
A new study by Ran Elkon and Ruth Ashery-Padan of Tel Aviv University and colleagues has revealed a previously unknown genetic risk factor for adult-onset macular degeneration (AMD) by combining a map of gene regulatory sites with disease-related loci. This discovery advances the knowledge of the main cause of visual loss in adults.
The findings were recently published in the journal PLOS Biology.
The leading cause of adult-onset macular degeneration (AMD) is due to a breakdown in the function of the retinal pigmented epithelium (RPE), a layer of tissue between the photoreceptors that detect light and the choriocapillaris that provides nourishment to the retina. Recognizing the significance of RPE in the development of AMD, the authors of the study focused on exploring the role of a transcription factor called LHX2. The team’s analysis of mouse mutants showed that LHX2 plays a crucial role in RPE development.
Knocking down LHX2 activity in RPE derived from human stem cells, they found that most affected genes were down-regulated, indicating that LHX2’s role was likely that of a transcriptional activator, binding to regulatory sites on the genome to increase the activity of other genes.
The authors found that one affected gene, called OTX2, collaborated with LHX2 to regulate many genes in the RPE. By mapping the genomic sites that OTX2 and LHX2 could bind to, they showed that 68% of those that bound LHX2 were also bound by OTX2 (864 sites in all), suggesting they likely work together to promote the activity of a large suite of genes involved in RPE development and function.
A common method for finding genes that may contribute to a disease is to perform a genome-wide association study (GWAS), which identifies genome sequence differences between individuals (termed single nucleotide polymorphisms, or SNPs) that co-occur with the disease. Numerous such studies have previously been done in AMD. However, a GWAS by itself cannot uncover a causal mechanism. Here, the authors compared their LHX2/OTX2 binding data to GWAS data in order to home in on variations that affected the binding of the transcription factors, and thus may contribute to disease.
One such binding site was located within the promoter region of a gene called TRPM1, which had been previously linked to AMD, and found that the sequence variant at that site altered the binding strength of LHX2; the so-called C version bound it more strongly than the T version, and activity of the TRPM1 gene was higher when the C allele was present instead of the T allele.
The results of the study indicate that the previously known increased risk of AMD from the variant identified in the GWAS was due to a reduction in the binding of the LHX2 transcription factor to the TRPM1 gene promoter, with a consequent reduction in the activity of this gene. The gene encodes a membrane ion channel, and previous studies have shown that mutations in the gene also cause visual impairment.
“Our study exemplifies how delineation of tissue-specific transcriptional regulators, their binding sites across the genome, and their downstream gene-regulatory networks can provide insights into a complex disease’s pathology,” the authors said.
Ashery-Padan adds, “The findings reveal a regulatory module consisting of LHX2 and OTX2 that controls the development and maintenance of the retinal pigmented epithelium, an important tissue of visual function. The genomic analyses further link the genomic regions bound by the two developmental factors to the genetics of the common, multifactorial blinding disease age-related macular degeneration (AMD).”
Reference: “The LHX2-OTX2 transcriptional regulatory module controls retinal pigmented epithelium differentiation and underlies genetic risk for age-related macular degeneration” by Mazal Cohen-Gulkar, Ahuvit David, Naama Messika-Gold, Mai Eshel, Shai Ovadia, Nitay Zuk-Bar, Maria Idelson, Yamit Cohen-Tayar, Benjamin Reubinoff, Tamar Ziv, Meir Shamay, Ran Elkon and Ruth Ashery-Padan, 17 January 2023, PLOS Biology.
DOI: 10.1371/journal.pbio.3001924



Be the first to comment on "Researchers Reveal New Genetic Risk Factors for Blindness"