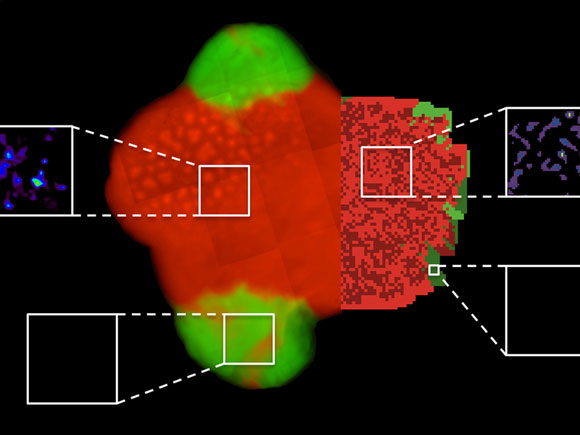
The experiment (bacterial colony and amino acid measurements by means of mass spectrometry imaging, left) corresponds to computer simulations (right): Cooperative bacteria are shown in red, non-cooperative bacteria are green. The opportunistic bacteria can only exist on the fringe of cooperating bacterial colonies. The white boxes show the concentrations of amino acids, which are high in the vicinity of cooperative bacteria (above). In contrast, virtually no amino acids were detectable in areas surrounding non-cooperative bacteria (below). © S. Pande / F. Kaftan, MPI for Chemical Ecology, S. Lang, Friedrich Schiller University Jena
New research shows that bacteria, which reciprocally exchange amino acids, stabilize their partnership on two-dimensional surfaces and limit the access of non-cooperating bacteria to the exchanged nutrients.
In natural microbial communities, different bacterial species often exchange nutrients by releasing amino acids and vitamins into their growth environment, thus feeding other bacterial cells. Even though the released nutrients are energetically costly to produce, bacteria benefit from the nutrients their bacterial partners provide in return. Hence, this process is a cooperative exchange of metabolites. Scientists at the Max Planck Institute for Chemical Ecology and the Friedrich Schiller University in Jena have shown that bacteria, which do not actively contribute to metabolite production, can be excluded from the cooperative benefits. The research team demonstrated that cooperative cross-feeding interactions that grow on two-dimensional surfaces are protected from being exploited by opportunistic, non-cooperating bacteria. Under these conditions, non-cooperating bacteria are spatially excluded from the exchanged amino acids. This protective effect probably stabilizes cooperative cross-feeding interactions in the long run.
The Research Group “Experimental Ecology and Evolution” headed by Christian Kost is investigating how cooperative interactions between organisms have evolved. In this context, the scientists study a special type of division of labor that is very common in nature, namely the reciprocal exchange of nutrients among unicellular bacteria. For these tiny organisms it is often advantageous to divide the labor of certain metabolic processes rather than performing all biochemical functions autonomously. Bacteria that engage in this cooperative exchange of nutrients can save a significant amount of energy.
Indeed, in a previous study, the researchers could already demonstrate that this division-of-metabolic-labor can positively affect bacterial growth. In the new study, they addressed the question of how such cooperative interactions can persist if non-cooperating bacteria consume amino acids without providing nutrients in return. The evolutionary disadvantage that results from cooperative cells could lead to a collapse of the cross-feeding interaction.
To experimentally verify this possibility, the scientists have monitored co-cultures of cooperating and non-cooperating bacteria. For this, they genetically engineered “cooperators” of two bacterial species that released increased amounts of certain amino acids into their environment. “As a matter of fact, non-cooperators grew better than cooperators in a well-mixed liquid medium, because, under these conditions, they had an unrestricted access to the amino acids in the medium. Their growth, however, was considerably reduced when placed on a two-dimensional surface,” said Kost, summarizing the results of the experiments. A more detailed analysis revealed that non-cooperating bacteria could only exist at the very fringe of colonies consisting of cooperating bacteria.
For their study, the scientists combined different methods and techniques. The basis formed a new research approach called “synthetic ecology”, in which certain mutations are rationally introduced into bacterial genomes. The resulting bacterial mutants are then co-cultured and their ecological interactions analyzed. In parallel, colleagues at the Friedrich Schiller University from the Department of Bioinformatics developed computer models to simulate these interactions. Finally, chemical analyses using mass spectrometric imaging was instrumental for visualizing the bacterial metabolites. Only the combination of microbiological methods with chemical-analytic approaches and computer simulations enabled the scientists to understand and elucidate this phenomenon.
“The fact that such a simple principle can effectively stabilize such a complex interaction suggests that similar phenomena may play important roles in natural bacterial communities,” Christian Kost states. After all, bacteria occur predominantly in so-called biofilms – these are surface-attached slime layers that consist of many bacterial species. Known examples include bacteria causing dental plaque or bacterial communities that are used in wastewater treatment plants. Moreover, biofilms are highly relevant for medical research: They not only play important roles in many infectious diseases by protecting bacterial pathogens from antibiotics or the patients’ immune responses, but are also highly problematic when colonizing and spreading on the surfaces of medical implants.
This new study has elucidated that cooperating bacteria form cell clusters and in this way exclude non-cooperating bacteria from their community. “The importance of this mechanism is due to the fact that no complicated or newly-evolved condition, such as the recognition of potential cooperation partners, needs to be fulfilled to effectively stabilize this long-term partnership. Two cooperating bacterial strains and a two-dimensional surface are sufficient for this protective effect to occur”, explains Kost.
The study raises many new exciting questions the researchers plan to address in the future. For example, they are interested in whether or not similar synergistic effects occur when more than two bacterial partners are involved. In their natural habitats, it is likely that more than two bacterial species participate in such cooperative interactions, leading to rather complex interaction networks. Moreover, amino acid-producing bacterial mutants were synthetically generated for this study. Whether also naturally evolved “cooperators” that occur in a habitat like soil show similar dynamics, remains to be verified. Given that bacteria frequently occur in biofilms, cooperative cross-feeding is probably much more widespread than previously thought. Understanding the factors and mechanisms that promote or inhibit bacterial growth could thus provide important clues on how to fight harmful bacteria or to better use beneficial ones.
Reference: “Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments” by Samay Pande, Filip Kaftan, Stefan Lang, Aleš Svatoš, Sebastian Germerodt and Christian Kost, 1 December 2015, The ISME Journal.
DOI:10.1038/ismej2015212








Bacteria does not have limbs to eat and fight.. Their enzymes produced by their DNA are only chemical weapons to combat with the environment. They will constantly change the DNA formula to survive in the environment. It is the fastest mutating organism. Any food it gets goes into the bacteria by Phagocytosis and its enzymes breaks down the carbonaceous material into amino acids. Naturally the production of enzymes and thereby amino acids (which are food for metabolism) would be in plenty. It is surprising that they don’t waste their excess amino acids and let it out to be used by other surrounding bacteria for their intake. These beneficial cooperative bacteria cluster into groups whereas those bacteria which do not have the excess enzymes do not contribute to mutually beneficial amino acid production and they are the non cooperating group. Nature will always go for the cooperative group which will be larger and the mutation in that direction will be fit for survival. Thank You.