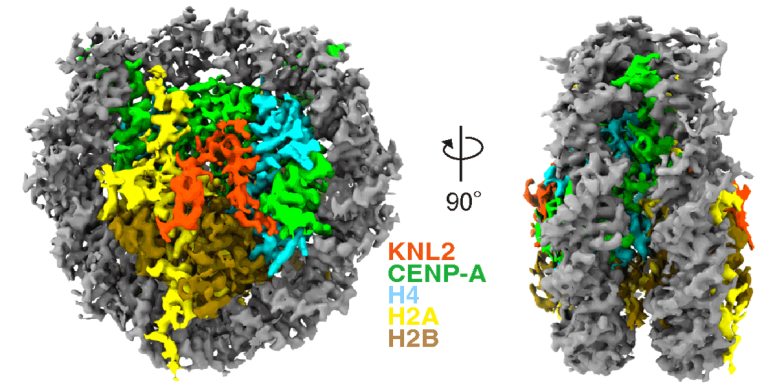
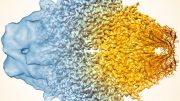
Cryo EM structure of the CENP-A nucleosome complex with KNL2. Credit: Jiang et al., EMBO J., 2023
Researchers led by Osaka University have used cryogenic electron microscopy analysis to reveal the structural change of the centromere at an atomic level during cell division.
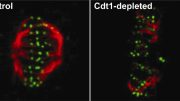
The genetic material inside cells is organized into structures called chromosomes. The centromere is essential for the correct division of the chromosomes via interaction with spindle microtubules when cells divide and grow. Now, a study by researchers at Osaka University has clarified the structure of the centromeric region in chicken cells using a technique known as cryogenic electron microscopy (cryo-EM).
Cryo-EM freezes samples quickly to preserve and stabilize them, and then images them using collisions with electrons to reveal their structure. A complex of proteins called the “kinetochore” forms at the centromeric region, and this is essential for cells to divide correctly. The researchers were able to clarify a structural change to the kinetochore at the atomic level using cryo-EM analysis.
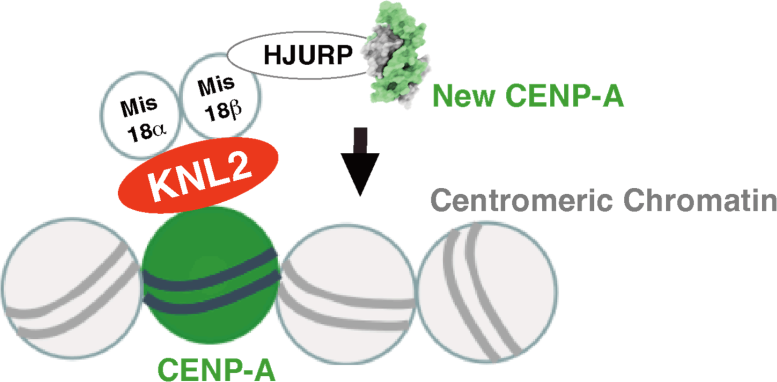
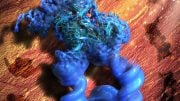
KNL2 binds to the CENP-A nucleosome during interphase and it contributes to new CENP-A deposition into centromeres via HJURP and the Mis18 complex. Credit: Original content by Tatsuo Fukagawa
When DNA is condensed into chromosomes, it is coiled around a core made of proteins called histones to form a structure known as a nucleosome. The nucleosomes in the centromeric region contain a variant histone protein called CENP-A, which specifies the location of the centromere. However, the mechanisms by which CENP-A is deposited at the centromeres to correctly define their location were unknown until now.
The research team showed that during mitosis (the process of cell division), a protein called CENP-C binds CENP-A and acts as a scaffold for other kinetochore proteins. However, during interphase (the time when the cell is not dividing), a different protein called KNL2 binds to centromeres instead. “KNL2 contains a CENP-C-like motif and is a component of the Mis18 complex, a licensing factor for new CENP-A deposition,” explains lead authors of the study Honghui Jiang and Mariko Ariyoshi.
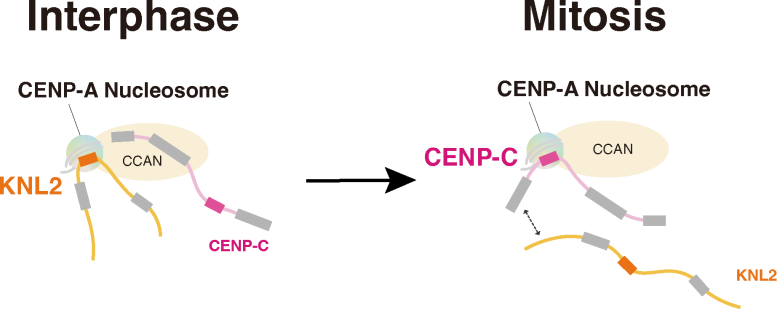
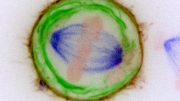
CENP-C excludes KNL2 from the CENP-A-KNL2 complex during mitosis. Credit: Original content by Tatsuo Fukagawa
The team further revealed that this interaction between KNL2 and the centromere is required for new deposition of CENP-A during interphase, which in turn helps maintain the correct location of the centromere. “We also showed that CENP-C is phosphorylated during mitosis, and phosphorylated CENP-C excludes KNL2 from the KNL2–CENP-A complex,” explains senior author Tatsuo Fukagawa. This suggests that KNL2 binds to CENP-A through interphase, maintaining the location of the centromere until a phosphate molecule becomes bound to CENP-C as the cells reach mitosis. Then, CENP-C preferentially binds to CENP-A, allowing the formation of the kinetochore for cell division.
These new insights into the structure of the centromeric region will prove invaluable in advancing knowledge of cell division and growth. Proteins involved in cell division and the kinetochore are targets for anti-cancer drugs; therefore, this work will also contribute to the design of novel drugs for diseases such as cancer.
Reference: “The cryo-EM structure of the CENP-A nucleosome in complex with ggKNL2” by Honghui Jiang, Mariko Ariyoshi, Tetsuya Hori, Reito Watanabe, Fumiaki Makino, Keiichi Namba and Tatsuo Fukagawa, 6 February 2023, EMBO Journal.
DOI: 10.15252/embj.2022111965
Funding: Japan Society for the Promotion of Science, Ministry of Education, Culture, Sports, Science and Technology, Japan Science and Technology Agency




Be the first to comment on "Cryogenic Electron Microscopy Reveals New Insights Into Centromere Structure"