Researchers have created a groundbreaking cellular map of a mammalian brain, detailing over 5,300 cell types in an adult mouse brain. This atlas, derived from extensive research, is a significant step in understanding brain function and evolution and holds promise for precision treatments of brain disorders. Credit: SciTechDaily.com
High-resolution atlas charts neural neighborhoods for more than 5,300 cell types.
Six years and 32 million cells later, scientists have created the first full cellular map of a mammalian brain. In a set of 10 papers in Nature today, a network of researchers unveiled an atlas cataloging the location and type of every cell in the adult mouse brain. Using advanced technologies that profile individual cells, the teams identified over 5,300 cell types – far more than known before – and pinpointed their locations within the brain’s intricate geography.
Unveiling the Brain’s Complex Architecture
Having a complete “parts list” of the brain will help accelerate efforts to unravel how it works, said Hongkui Zeng, Ph.D., Executive Vice President and Director of the Allen Institute for Brain Science.
“This is a landmark achievement that really opens the door for the next stage of investigations of the brain’s function, development and evolution, akin to the reference genomes for studying gene function and genomic evolution,” said Zeng, who led one of the studies. “My colleagues said that the 5,000 cell types we identified will keep neuroscientists busy for the next 20 years trying to figure out what these cell types do and how they change in disease.”
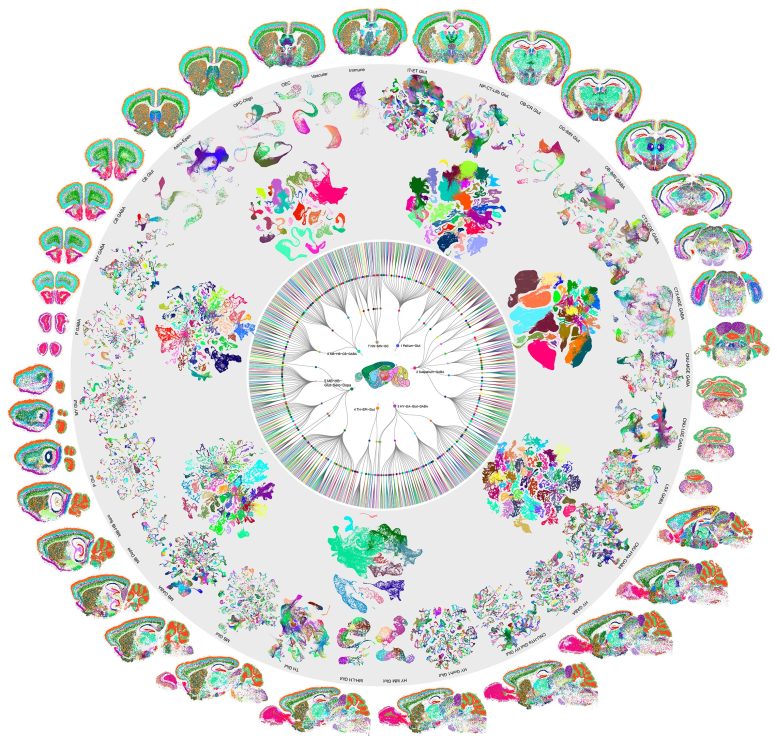
Detailed classification and distribution of cell types in the entire mouse brain based on the expression of their genes. More than 5000 cell types have been identified and can be grouped based on their similarity to each other. Each of the groups was plotted as a UMAP to highlight the relationships between cell types in specific groups and each group can be assigned to specific anatomical location. Credit: Allen Institute
The collective work is a capstone for the National Institutes of Health’s BRAIN Initiative Cell Census Network, or BICCN. Hundreds of researchers contributed to the project, which was funded by the NIH’s Brain Research Through Advancing Innovative Neurotechnologies® (BRAIN) Initiative, or The BRAIN Initiative®.
“Where we previously stood in darkness, this milestone achievement shines a bright light, giving researchers access to the location, function, and pathways between cell types and cell groups in a way we couldn’t imagine previously,” said John Ngai, Ph.D., Director of the NIH BRAIN Initiative. “This product is a testament to the power of this unprecedented, cross-cutting collaboration and paves our path for more precision brain treatments.”
Linking Genetics to Brain Geography
By combining single-cell RNA sequencing with spatial transcriptomics—methods for determining which genes are expressed in individual cells and where those cells are located—Zeng and her collaborators revealed the brain’s astonishing complexity and diversity.
One of the atlas’s major revelations is the deep connection between a cell’s genetic identity and its spatial position, Zeng said. This relationship underscores how location shapes function, offering clues into the evolutionary history and intricate interactions of different brain regions.
“We’re seeing the building blocks of the brain’s circuits,” she said. “The brain’s organization likely reflects its evolutionary history.”
One intriguing finding is the distinct cellular organization between the lower (“ventral”) versus the upper (“dorsal”) parts of the brain. While the ancient ventral part features a mosaic of interrelated cells, the more recent dorsal part contains fewer but highly divergent cell types. This distinction could be a key to deciphering how different brain regions evolved unique roles, for example, the ventral part for basic survival and the dorsal part for adaptation, Zeng said.
Unlocking the Secrets of Cell Communication
The researchers also found that transcription factors – proteins that regulate gene activity – comprise a ‘code’ that specifies a cell’s identity.
The atlas also uncovered how brain cells talk to each other via a diverse cast of signaling molecules, which carry messages from cell to cell. That diversity enables complex interactions between different cell types.
The strong alignment across independently collected genomic, epigenomic, and spatial datasets provides high confidence that this atlas maps more than just cell identities – it captures the true organizational blueprints underlying mammalian brain development, Zeng said.
The Path Forward: Applications and Extensions
Looking ahead, the atlas can serve as a model for similar mappings in the brains of other species—namely our own. That work is already underway.
It also provides a guide to genetically target specific cell types, enabling tools to study specific functions and disease. This could pave the way for precision treatments, Zeng said.
“We know that many diseases originate in specific parts of the brain, and probably in specific cell types,” she said. “With this map in hand, we can gain a more precise view of the dysfunction of disease and then create genetic or pharmacologic tools to target those specific cell types, to achieve greater efficacy and minimal side effects.”
Contributions to Spinal Cord Research
Allen Institute scientists also co-led a study to create a detailed map of the neurons that connect the brain to the spinal cord, enabling movement and sensory modulation. In this study, a team led by Zhigang He, Ph.D., and Carla Winter, M.D., Ph.D., of Harvard provide the most in-depth characterization of these spinal-projecting neurons (SPNs) to date. By integrating the molecular identities and locations of these neurons into one atlas, scientists gain insight into how this intricate network controls function and movement. “And by having a baseline map of these cell types, we can now study how spinal cord injuries or stroke alter them and hopefully develop targeted therapies,” said Winter.
Allen Institute scientists contributed to five other studies, including:
- A spatial atlas of cell types in the whole mouse brain. In this study, led by Xiaowei Zhuang, Ph.D., of Harvard, scientists used spatially resolved transcriptomic profiling of >1,100 genes to reveal the spatial organization of >5,000 transcriptionally distinct clusters across the entire mouse brain. Registration of the cell atlas to the Allen Common Coordinate Framework allows quantification of cell type composition and organization in each brain region. The high-resolution spatial map reveals cell-cell interactions and molecular underpinnings between hundreds of cell-type pairs.
- A comparison of gene regulatory programs across different species, including humans. In this study, researchers analyzed certain regions of DNA that act like switches, turning genes on or off and controlling a cell’s identity. The team found that so-called jumping genes—DNA sequences that can flit about the genome—make up the bulk of human-specific “switches” in the neocortex. As these same regions can also be involved in neurodegenerative diseases, further study could point the way to new therapies, the authors said. “This data is a gold mine for geneticists who can now start to uncover the molecular basis of complex traits like schizophrenia,” said Bing Ren, Ph.D., of UCSD, who co-led the study with the Salk Institute’s Joseph Ecker, Ph.D.
References:
“Molecularly defined and spatially resolved cell atlas of the whole mouse brain” by Meng Zhang, Xingjie Pan, Won Jung, Aaron R. Halpern, Stephen W. Eichhorn, Zhiyun Lei, Limor Cohen, Kimberly A. Smith, Bosiljka Tasic, Zizhen Yao, Hongkui Zeng and Xiaowei Zhuang, 13 December 2023, Nature.
DOI: 10.1038/s41586-023-06808-9
“Conserved and divergent gene regulatory programs of the mammalian neocortex” by Nathan R. Zemke, Ethan J. Armand, Wenliang Wang, Seoyeon Lee, Jingtian Zhou, Yang Eric Li, Hanqing Liu, Wei Tian, Joseph R. Nery, Rosa G. Castanon, Anna Bartlett, Julia K. Osteen, Daofeng Li, Xiaoyu Zhuo, Vincent Xu, Lei Chang, Keyi Dong, Hannah S. Indralingam, Jonathan A. Rink, Yang Xie, Michael Miller, Fenna M. Krienen, Qiangge Zhang, Naz Taskin, Jonathan Ting, Guoping Feng, Steven A. McCarroll, Edward M. Callaway, Ting Wang, Ed S. Lein, M. Margarita Behrens, Joseph R. Ecker and Bing Ren, 13 December 2023, Nature.
DOI: 10.1038/s41586-023-06819-6







So the microfluidics have been enlarged in the 10x Chromium chips, and 10X says they only have “vaidated” cells with 35 micron diameter. Neurons are way bigger and even though they can deform are most likely being shredded. I’ve noticed that some of these paper de-emphasize the scRNA-seq in the methods and refer you to another paper, this is wise. It’s also hard to pretend that you didnt know either. SMH