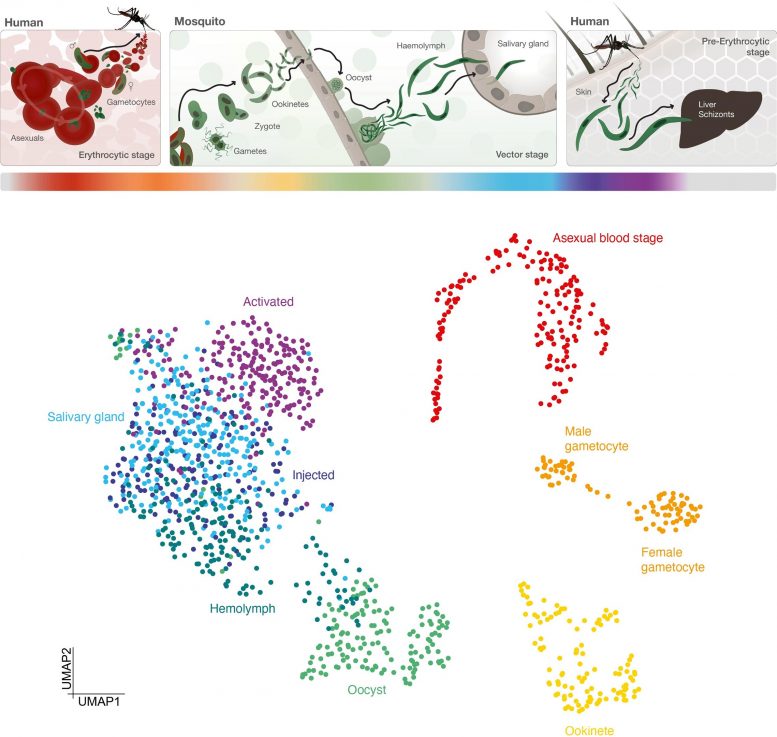
Transmission cycle of malaria parasites between human host and mosquito vector (upper) and map of gene activity of parasite cells at different stages of the cycle (bottom). Each color represents a stage of the life cycle. Credit: Real et al.
Researchers have mapped in fine detail the genetic changes malaria parasites go through as they prepare to infect people.
The atlas maps the malaria parasite Plasmodium falciparum in unprecedented cellular detail as it develops inside a mosquito and prepares to infect humans through a bite. This detailed investigation could lead to new ways to block key stages in the parasite’s development and prevent transmission through future drugs or vaccines.
Mosquitoes are increasingly resistant to pesticides, and the parasite that causes malaria is also becoming increasingly resistant to antimalarial drugs. This has created an urgent need for new ways to fight malaria, which in 2019 caused an estimated 229 million cases and 409,000 deaths, most of which were young children in sub-Saharan Africa.
To reinvigorate efforts in drug or vaccine discovery, a team from the lab of Professor Jake Baum at Imperial College London and the lab of Dr. Mara Lawniczak from the Wellcome Sanger Institute have examined the human malaria parasite Plasmodium falciparum in unprecedented detail. Their results are published today (May 27, 2021) in Nature Communications.
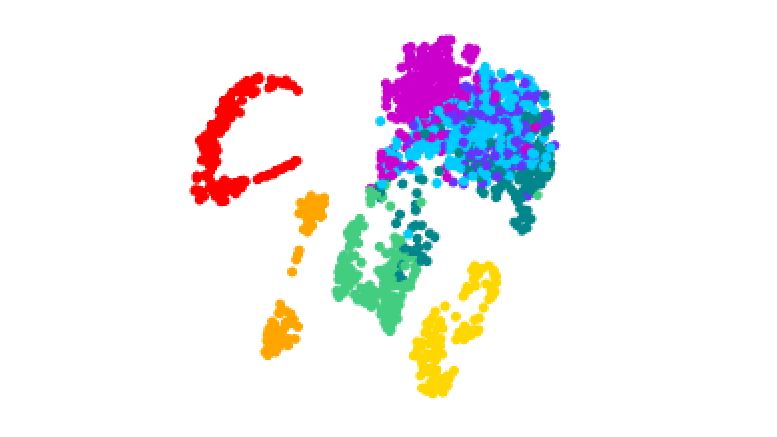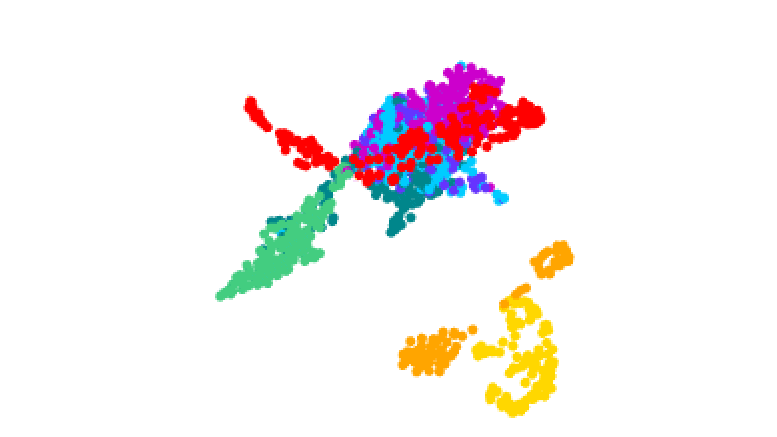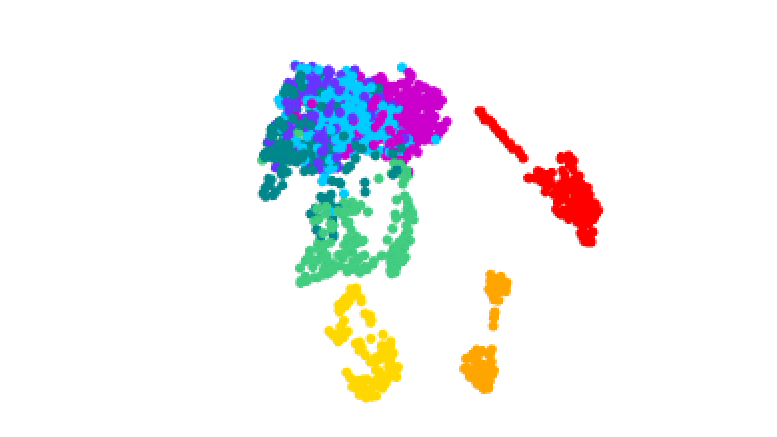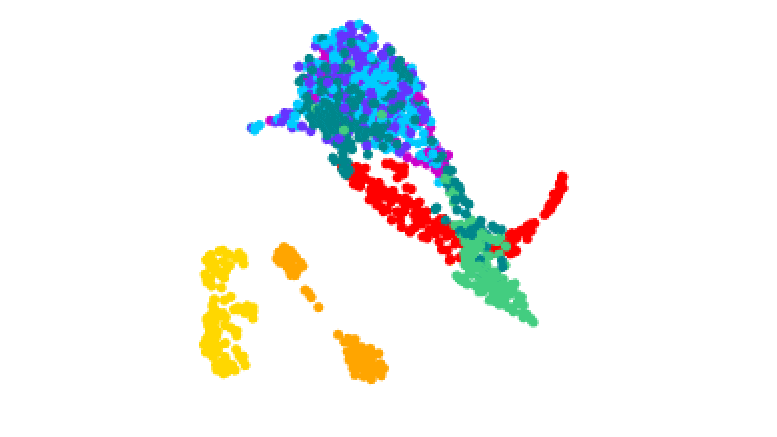
3D plot of gene activity in human malaria parasites at different stages of the life cycle. Credit: Real et al.
P. falciparum develops in the midgut of a mosquito before traveling to the mosquito’s saliva glands, ready to infect a human when the bug bites. During these phases, the parasite goes through many stages important for its development and ability to transmit, including changing into different forms.
The team tracked how these stages were controlled by analyzing the activity of genes throughout the process. They isolated the different forms of the parasite and produced 1467 ‘transcriptomes’ – maps of which genes in single cells are turned on or off during the different stages.
When genes are turned on, they instruct the cell to make different proteins and drive developmental changes, such as causing the parasite to exit the midgut and colonize the salivary gland of the mosquito, or to travel through human cells to reach the liver, where the parasite prepares to invade more human cells.
Knowing how these processes work in detail at the cellular level reveals to researchers new targets that could be blocked to stop development, preventing transmission of the parasite.
Dr. Eliana Real, from the Department of Life Sciences at Imperial, said: “Being directly based on the human-infective parasite, our new data have clear implications for malaria control, which has an increasing focus on transmission blocking strategies both in terms of drugs that kill the parasite as it moves between stages and protective vaccines. Understanding how parasites behave transcriptionally within the mosquito vector provides a foundation from which new strategies will surely arise.”
As well as surveying the whole transmission cycle of the parasite, the team focused on what is called the sporozoite stage: the form released into the human skin during a mosquito bite. They sorted parasites from within the mosquito during their development, and isolated sporozoites after an infectious bite as they interact with human skin cells. In doing so, they were able to find specific patterns of gene expression that define each of the critical stages in these processes.
Dr. Virginia Howick, previously from the Wellcome Sanger Institute and now based at the University of Glasgow, said: “This fine granularity enables us to trace sporozoite developmental processes and to propose new mechanistic targets essential for each step and future vaccine targets for blocking malaria infection.”
The team were also able to compare their data with a similar set from the related parasite Plasmodium berghei, a rodent malaria parasite that is often used as a model for studying malaria disease in the lab. This showed which genes are common between species, and which are specific to the human version of the parasite.
Dr. Farah Dahalan, from the Department of Life Sciences at Imperial, said: “This level of gene surveillance at the individual parasite level throughout its life cycle will provide an invaluable resource for researchers to discover previously unexplored elements of Plasmodium cell biology, comparative Plasmodium species biology and the development of control methods that target particular pathways or lay the foundations for improving vaccines.”
The researchers have made all their data available on an interactive website, where the transcriptional profile of any gene across any stage of the Plasmodium life cycle can be easily and freely viewed.
Reference: “A single-cell atlas of Plasmodium falciparum transmission through the mosquito” by Eliana Real, Virginia M. Howick, Farah A. Dahalan, Kathrin Witmer, Juliana Cudini, Clare Andradi-Brown, Joshua Blight, Mira S. Davidson, Sunil Kumar Dogga, Adam J. Reid, Jake Baum and Mara K. N. Lawniczak, 27 May 2021, Nature Communications.
DOI: 10.1038/s41467-021-23434-z
The research was funded by Wellcome, the Bill & Melinda Gates Foundation, and the Royal Society.






Be the first to comment on "Atlas of Malaria Parasite Genetic Activity Provides New Targets for Drugs and Vaccines"