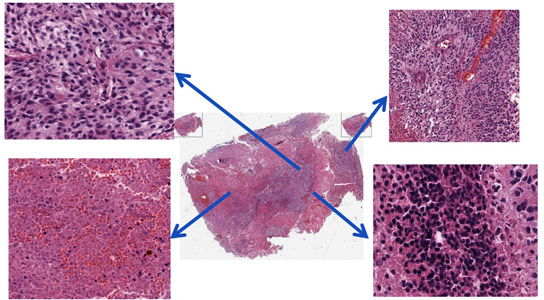
A tumor’s organizational complexity is revealed. The center image is a whole-slide image of a Glioblastoma Multiforme tumor. The arrows indicate enlarged, distinct regions. Berkeley Lab scientists have developed an automated way to analyze large sets of tumor images. Credit: Lawrence Berkeley National Laboratory
Scientists at Berkeley Lab have developed an algorithm and a computational pipeline that combs through large sets of images and identifies tumor subtypes, helping shed light on whether tumor subtype can predict the effectiveness of therapies and allowing scientists to learn more about the genetic and molecular mechanisms that control tumor signatures.
How’s this for big data: A whole-slide image of a tumor section can be ten billion pixels. There can be thousands of such images in the tumor cohorts maintained by The Cancer Genome Atlas project, which are collected from a large pool of patients.
The images are a potential treasure trove for the emerging field of precision medicine. Hidden in those billions of pixels is a story of how tumor cells organize themselves, the molecular networks that influence these structural traits, and what it all means for patients. Unfortunately, culling this information from numerous images is difficult. That’s because no two tumors are alike, and there are myriad technical variations in how samples are prepared.
This analysis could soon get much easier. Berkeley Lab scientists have developed an algorithm and a computational pipeline that combs through large sets of images and identifies tumor subtypes. It also identifies heterogeneity, or the extent to which a tumor comprises different organizational structures. The pipeline then uses clinical data to rank cellular signatures that are predictive of patient outcome. It also uses large-scale genomic data to identify molecular correlates of each subtype.
The resulting information will help scientists learn more about the genetic and molecular mechanisms that control tumor signatures. It will also shed light on whether tumor subtype can predict the effectiveness of therapies.
“Our goals are to identify morphometric and architectural traits that can be predictive of a therapy. We’d also like to learn about the molecular signatures that lead to architectural aberrations,” says Bahram Parvin of Berkeley Lab’s Life Sciences Division. The development of the core computational module and the pipeline were led by Hang Chang and Gerald Fontenay, respectively, in Parvin’s Lab in the Life Sciences Division.
The core computational module works by extracting each cell from an image, and then profiling properties of each cell such as size, shape, and organization. In this way, the telltale characteristics of a specific tumor subtype are gleaned from a large cohort of images.
As recently reported, the scientists validated their pipeline by applying it to 377 whole-slide images from 146 patients who have an aggressive brain cancer called Glioblastoma Multiforme. The pipeline identified several tumor subtypes based on a range of cellular profiles. It also determined whether each subtype is predictive of a patient’s response to alternative therapy. Although the pipeline was developed in a high-performance computer language, it is compute-intensive and requires extensive use of the Lawrencium cluster operated by Berkeley Lab’s IT Division.
The scientists also created an online repository (at tcga.lbl.gov) for these images, which also includes images of low-grade glial and kidney renal carcinoma tumor sections. The website allows for Google-map style zooming and panning of the tissue sections. The scientists next hope to layer more information onto the images, in addition to cellular structure, to provide a broader representation of the tumors’ characteristics and interactions between different components of tumor histology.
Reference: “Invariant Delineation of Nuclear Architecture in Glioblastoma Multiforme from The Cancer Genome Atlas Cohort” by Hang Chang, Ju Han, Alexander Borowsky, Leandro Loss, Joe W Gray, Paul T Spellman, Bahram Parvin, 4 December 2012, IEEE Transactions on Medical Imaging.
DOI: tmi.2012.2231420








Be the first to comment on "Berkeley Lab Develops a Computational Pipeline to Analyze Tumor Images"