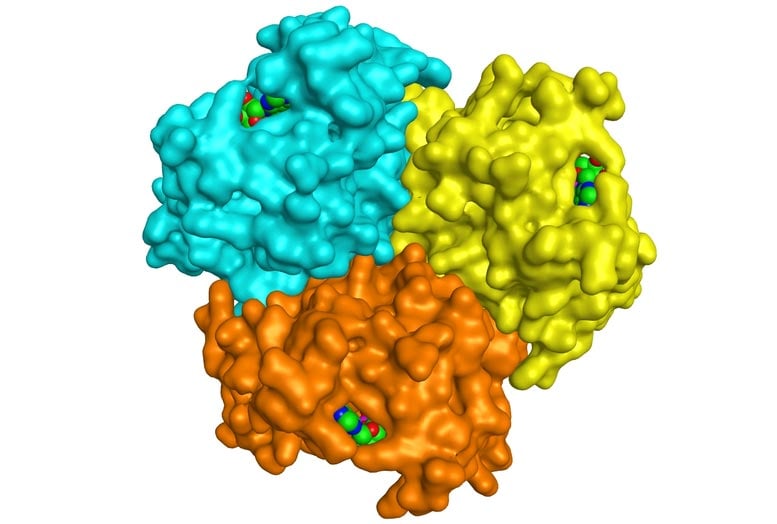
Trimeric structure of an ancient enzyme from Odin archaea, with substrate molecules shown in green. Credit: Magnus Wolf-Watz
“The distinction between the past, present, and future is only a stubbornly persistent illusion,” Albert Einstein wrote. Perhaps this is nowhere more evident than in protein evolution, where past and present versions of the same enzyme exist in different species today, with implications for future enzyme design. Now, researchers have used evolutionary “time travel” to learn how an enzyme evolved over time, from one of Earth’s most ancient organisms to modern-day humans.
The researchers will present their results today at the fall meeting of the American Chemical Society (ACS). ACS Fall 2021 is a hybrid meeting being held virtually and in-person August 22-26, and on-demand content will be available August 30-September 30. The meeting features more than 7,000 presentations on a wide range of science topics.
“If a person lives in present-day Rome, they might want to learn about ancient Rome to better understand who they are,” says Magnus Wolf-Watz, Ph.D., the project’s principal investigator. “In the same way, we can look backward in time at more ancient forms of enzymes to understand how the proteins are working today and how we might engineer new versions in the future.”
Wolf-Watz, who is at Umeå University in Sweden, looked back some 2 to 3 billion years to primitive organisms known as archaea. These single-celled life forms, which still exist today, have characteristics of both prokaryotes (bacteria, which lack a cell nucleus) and eukaryotes (organisms like plants, animals and fungi that have a nucleus in their cells). A branch of archaea known as the Asgard phylum, discovered in 2015, comprises the closest known ancestors to eukaryotic cells. Four types of Asgard archaea have been identified, including Odin archaea, found in hydrothermal vents deep in the Atlantic Ocean.
Odin archaea have an enzyme called adenylate kinase (AK), which is also found in prokaryotes and eukaryotes. Wolf-Watz previously studied two human types of this enzyme, AK1 and AK3. Both are important in maintaining the energy balance in cells, but AK1 is in the cytoplasm, where it transfers a phosphate group from adenosine triphosphate (ATP, the main energy carrier in cells) to adenosine monophosphate (AMP). In contrast, AK3 resides within mitochondria, where it transfers a phosphate group from guanosine triphosphate (GTP, a molecule similar to ATP but with distinct roles) to AMP.
Wolf-Watz’s team used X-ray crystallization and nuclear magnetic resonance spectroscopy to study the structures of AK1 and AK3, finding that although the enzymes are very similar, they have a subtle difference in a short loop region that causes AK1 to prefer ATP and AK3 to prefer GTP. “Now we can take any AK enzyme, look at the structure of that loop, and predict whether it’s going to use ATP or GTP,” Wolf-Watz says. The next step was to examine a more ancient version of an AK enzyme –– from Odin archaea –– to learn how AK1 and AK3 evolved to prefer different nucleotide substrates.
The researchers purified the archaea AK and determined its structure. They found that the loop important for discriminating ATP and GTP is much longer in the archaeal enzyme, and it has chemical groups that can bind either nucleotide. “What we found is an early ancestor of the human AKs that contains two capacities –– it can use both ATP and GTP,” Wolf-Watz says. “During the course of evolution, it became specialized to become specific for one or the other, depending on the cellular compartment where it resides.” The archaea AK can actually use all naturally occurring nucleotide triphosphates (NTPs). “We’ve discovered a universal NTP binding motif that could be a building block for the future design of novel enzymes,” Wolf-Watz says.
The archaeal AK contains three copies of the enzyme (known as a trimer) that bind to each other through a helical structure. In human AKs, a mutation in this region makes the enzyme copies unable to stick to each other. The human enzymes, which function independently, are almost 1,000-fold more active. The trimer could have been more stable in the extreme environment of hydrothermal vents, but the human enzymes might have traded this thermostability for higher activity, which is important in a cooler environment, Wolf-Watz says.
Next, the researchers want to engineer novel enzymes that could be useful in organic synthesis or drug development. They also want to examine other enzymes from Odin archaea and study how they might have evolved over the eons. “We studied one enzyme and made this fantastic discovery,” Wolf-Watz says. “Of course, there’s more to find. It’s like we’re digging through a treasure chest.”
A recorded media briefing on this topic will be posted Wednesday, August 25 at 9 a.m. Eastern time at www.acs.org/acsfall2021briefings.
The researchers acknowledge support and funding from the Swedish Research Council, the Kempe Foundations and the Carl Trygger Foundation.
Title
Evolutionary origins of enzymatic specificity and dynamics
Abstract
“Time-travel” is a concept that carry significant potential to decode previously unknown aspects of protein function. Archeological “backwards-looking” can be performed with evolutionary analysis, while a glimpse into the future can be obtained from directed evolution and enzyme design. Here, I will present our evolutionary approach that is centered on the enzyme adenylate kinase (AK) isolated from organisms from all three kingdoms of life; bacteria, archaea and eukarya. For the archaeal organism we have selected Odinarchaeota, a member of the recently discovered Asgard archaeal family that is believed to be the closest evolutionary ancestor to modern eukaryotic organisms. Comparative structural and functional analysis between AK from these three domains has enabled us to uncover novel principles in enzymatic catalysis. I will specifically present findings for the evolutionary origin of nucleoside triphosphate (NTP) specificity, but also for the evolutionary origin of large-scale conformational dynamics. The findings have been possible through an integrative structural biology approach where we utilize state-of-the-art quantitative 19F NMR spectroscopy (relaxation dispersions) for determination of microscopic rate-constants for conformational dynamics of a large (69 kDa) assembly.








Be the first to comment on "Evolutionary “Time Travel” Reveals Enzyme’s Origins, From One of Earth’s Most Ancient Organisms"