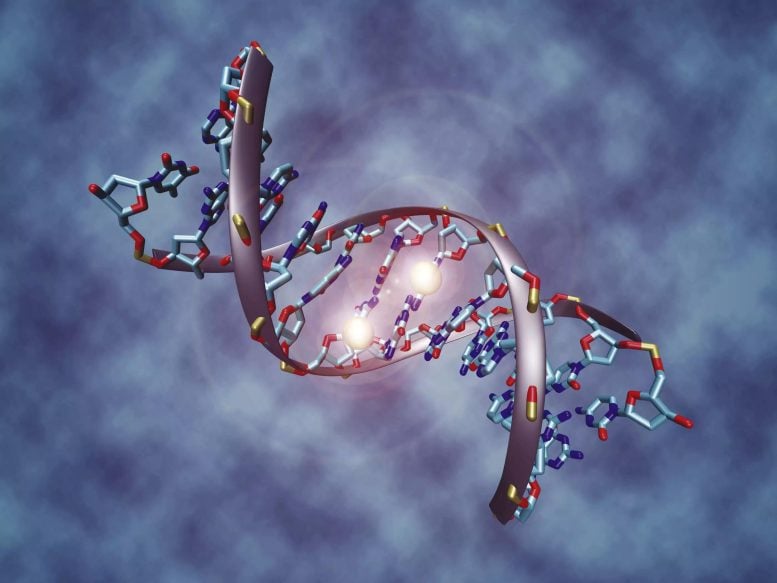
Methylated bases of DNA – highlighted with bright dots in this image – deactivate a gene. The methylation pattern reveals if chemotherapy is effective against a tumor, here visible as a bright spot in the brain section. Credit: MPI for Informatics
A computational epigenetics research group at the Max Planck Institute for Informatics in Saarbrücken is using software programs to rummage through the genomes of cancer patients in search of suspicious methylation patterns. They use these patterns in clinical diagnosis as biomarkers and combined with ingenious algorithms and statistical processes, researchers have developed an epigenetic biomarker for malignant glioblastoma.
The relatively young research field of epigenetics is the talk of the town. Many scientists expect the research on biochemical modifications beyond the actual DNA strand to lead to huge progress in the understanding of the regulation of gene activity in the years to come. Just how promising the results of epigenetic research are in terms of concrete medical applications is demonstrated by the work of Thomas Lengauer and Christoph Bock from the Max Planck Institute for Informatics in Saarbrücken. With the help of computers, they trawl through the genomes of cancer patients in search for suspect structures, and develop fast and simple new tools for improving cancer diagnosis in hospitals.
Although the human body consists of a scarcely conceivable 10 to 100 billion cells, it only takes a few diseased cells to cause cancer. Minor damage in the DNA as a result of exposure to UV light or tobacco smoke can switch off a cell’s natural growth limits: the cell then starts to divide uncontrollably and, in the worst-case scenario, overgrow healthy tissue in the form of tumors, which eventually destroy vital organs.
It was long believed that only changes in the DNA itself play a crucial role in the emergence of cancer. However, it is now clear that the biochemical “coat” of the DNA strand also has an important role to play, as a cell’s genetic material is modified by a large number of chemical attachments. DNA methylation, in particular, plays a central role in gene regulation: small hydrocarbon attachments thus decide whether a gene is “active” or “silenced,” namely whether it can be read off or not.
Defects in DNA methylation result in altered gene activity in the cell which can contribute to tumor formation. In addition, the methylation patterns of tumor cells differ clearly from those of healthy tissue cells. “This is the precise point where our research begins,” says Thomas Lengauer from the Max Planck Institute for Informatics in Saarbrücken, who, together with Christoph Bock, leads the Computational Epigenetics Research Group in Professor Lengauer’s Department of Computational Biology and Applied Algorithmics.
The scientists rummage through vast collections of genetic data for suspicious methylation patterns using software programs they develop themselves. “Many of these patterns only arise in very specific types of cancer, so we can use them in clinical diagnosis as biomarkers, that is to say as indicators for the corresponding form of the disease,” says Thomas Lengauer. The scientists rely on close cooperation with hospitals and biotechnology laboratories for their work. The tissue samples from cancer patients are processed in the laboratory and the genetic material they contain is cut into numerous small snippets. The solution is then processed using microarrays (DNA chips) or using new-generation sequencing processes. “Eventually you get a kind of map of the epigenome, which comprises all of the biochemical markings layered on top the actual DNA sequence,” explains Thomas Lengauer.
The subsequent mathematical analyses by the Saarbrücken-based researchers are carried out exclusively in the computer. Ingenious algorithms and statistical processes screen the sea of epigenome data for patterns that only arise in certain forms of cancer and are absent in healthy patients – a task that requires a lot of careful programming. Because, unlike the genome, which is almost identical in all of the body’s cells, the DNA’s chemical coat is a real chameleon. “There are around 200 different types of tissue in the human body, each with different epigenomes. These also change with advancing age and in the presence of disease,” says Thomas Lengauer. “We have to take all of these variations into account in our programs to ensure that only differences relevant to cancer are considered important.”
And all of this effort is proving worthwhile. Working in collaboration with the Universitätsklinikum Bonn, the scientists in Saarbrücken developed an epigenetic biomarker for malignant glioblastoma, the most common form of malignant brain tumor. “Chemotherapy is only effective in around one quarter of affected patients,” explains Christoph Bock, who also leads a research group at the Research Center for Molecular Medicine in Vienna. “In these patients, a particular gene called MGMT is methylated, that is silenced. In its active state, this gene controls a repair mechanism in the cancer cells. In patients with active MGMT, the DNA damage arising during chemotherapy, which causes diseased cells to die, would be reversed and the treatment fails.” With the help of the new biomarker, the hospital can now identify in advance of treatment those patients for whom debilitating chemotherapy is actually worthwhile. The researchers subsequently systematized the time-consuming manual procedure for the identification of this biomarker and developed a corresponding software program. Using this software, other innovative biomarkers for cancer can now be identified considerably faster.
The researchers’ latest success has also opened up completely new paths in clinical cancer diagnosis. In a major international project headed by Manel Esteller from the Bellvitge Biomedical Research Institute in Barcelona, Spain, over 1,600 human tissue samples were analyzed. The methylation pattern was sampled at around 1,500 characteristic places in each analyzed genome and the resulting data analyzed by computer at the MPI in Saarbrücken. The results of the analysis are very promising. Even more significant, however, is the fact that, based on the methylation patterns, the researchers were able to classify tumors that belong to the “cancers of unknown primary origin” group, known as CUPs.
“Imagine that a patient comes to the doctor complaining about pains in his liver,” says Christoph Bock. “The doctor then establishes that numerous metastases originating from an unknown primary tumor have already formed in the liver.” When such malignant cancer cells of unknown origin are found in the body, the frantic search for the primary tumor begins. In around 25 percent of cases, this search is unsuccessful. Because cancer cells frequently degenerate outside their original tissue, without knowing the origin of the primary tumor, it is almost impossible to establish what kind of cancer is involved and this, in turn, significantly reduces the patient’s chances of a successful cure. “Through epigenome analysis we were able to assign 70 percent of the CUPs in our samples definitively to one type of cancer,” says Christoph Bock. “Therefore, analytical tools like those we have developed for glioblastoma can improve the prognosis for these patients because the hospital then can select the treatment best suited to their condition.”
In the meantime, the potential offered by epigenome mapping has also been recognized at an international level. Various major projects are being coordinated under the aegis of the International Epigenome Consortium (IHEC). The general aim is to completely map the epigenomes of at least 1,000 biologically and medically significant cell types and cell states. The individual projects within the IHEC focus on different issues. Whereas, for example, the US institutes are aiming to create reference profiles for as many healthy human cell types as possible, the EU-funded BLUEPRINT project is focusing on the cells of the blood and immune system. “The approach adopted by BLUEPRINT is extremely application-oriented as many clinical diagnoses are based on the analysis of blood samples,” says Christoph Bock. In addition to around 60 healthy cell types in the blood, it is also intended to map over 60 forms of leukemia. The MPI in Saarbrücken is not the only Max Planck Institute involved in IHEC: the MPI of Immunobiology and Epigenetics in Freiburg and the MPI for Molecular Genetics in Berlin are also participating in this project.
“Thanks to bioinformatics, our knowledge in the field of epigenetics will proliferate in the decade to come,” says Thomas Lengauer. The professor from Saarbrücken continues: “In the future, epigenetics will form the backbone of the explanation as to how cells control themselves – a central question in biology. With the help of bioinformatics-related methods it will be possible to discover many processes in the cell nucleus that are still completely unknown, in particular the regulation of gene activity.”
Data management is the first task in the long to-do list. “The wave of data that is now approaching is nothing short of revolutionary,” says Thomas Lengauer. “Here in Saarbrücken, the servers are bursting at the seams, so much so that we are currently considering moving our data, in part at least, to the cloud.” Obviously, the epigenome maps must be stored in a network, be easily accessible and be made usable with the help of suitable tools and search engines. “The alternative now being given serious consideration would mean that laboratories and clinics would have to re-sequence stored samples each time they need them, simply because there is no space to store the data,” says Lengauer. “A highly remote solution – for computer scientists at least.”
Although Thomas Lengauer regards epigenome analysis as playing a crucial role in the attainment of rapid progress in cancer diagnosis in the near future, he plays down expectations with regard to the development of new drugs. “Many scientists point to the potential of future drugs that can repair defects in the epigenome of diseased cells. I tend to be more cautious in this regard. Such targeted interventions involve significant risks, not least because little or nothing is currently known about the highly-complex gene regulation mechanisms being manipulated here.”







Be the first to comment on "Researchers Use Bioinformatics and Epigenetics to Aid Cancer Research"