The development of mRNA knock-down technologies for use in vertebrate organisms such as zebrafish has been limited. CRISPR-RfxCas13d has been identified as an effective method for the systematic study of gene function in vivo during embryogenesis across a range of animal species. Credit: Bazzini Lab, Stowers Institute
CRISPR is a gene-editing tool that has enabled researchers to study the function of different genes in model organisms. Scientists at the Stowers Institute for Medical Research in Kansas City, Missouri, and the Andalusian Center of Developmental Biology at Pablo de Olavide University in Seville, Spain, have now harnessed this powerful technology to target gene messages in animal model embryos to gain a better understanding of the genetic programs that direct early stages of vertebrate development.
In a report published online today (August 7, 2020) in Developmental Cell, the Stowers team, led by Assistant Investigator Ariel Bazzini, Ph.D., and collaborators show that the technology, called CRISPR-Cas13, is able to target RNA — DNA’s chemical cousin that carries messages needed to construct proteins — in embryonic animal models in a specific and systematic manner, allowing researchers to study the role of RNA in the earliest hours of development. The researchers show that the CRISPR-Cas13 method is effective in zebrafish, killifish, medaka, and mouse embryos, and thus could be used to explore early developmental genetic programs in a broad range of animal species.
Since its discovery, scientists have been using the much-lauded gene editing tool CRISPR to alter the DNA of model organisms and uncover the functions of thousands of genes. Now, researchers at the Stowers Institute for Medical Research in Kansas City, Missouri, and the Andalusian Center of Developmental Biology at Pablo de Olavide University in Seville, Spain, have harnessed the technology to target gene messages (messenger RNA) involved in early vertebrate development.
By disrupting gene messages (RNA) instead of the underlying genes (DNA), researchers can study genes that might previously have been difficult or impossible to manipulate because they were essential to life or involved in a critical stage of biological development. This approach also allows targeting of maternally-contributed gene RNAs, which are deposited in the egg to kick off the earliest genetic programs.
The study, which appears online August 7, 2020, in the journal Developmental Cell, establishes the use of CRISPR-Cas technology to target RNA in embryonic animal models in a specific and systematic manner. The findings demonstrate the technique can be applied to a broad range of aquatic and terrestrial models including zebrafish, medaka, killifish, and mice.
“The exciting thing about this study is not just what we found, but what we can do,” says Ariel Bazzini, Ph.D., an assistant investigator at the Stowers Institute and co-leader of the study. “We still don’t understand how genes jumpstart the earliest stages of development. Now we can find out by targeting their RNA messages, one by one.”
“We are also very excited about the low cost of the technique,” Bazzini says. “Any lab working with zebrafish or other animal embryos could use this method. Indeed, we have already distributed the reagents and protocol to several labs around the world.”
Before development even begins, egg must first meet sperm. The resulting embryo carries half the genes from the mother and half from the father. In addition to its genome, the embryo has components such as RNA and proteins provided by the mother.
The development of mRNA knock-down technologies for use in vertebrate organisms such as zebrafish has been limited. CRISPR-RfxCas13d has been identified as an effective method for the systematic study of gene function in vivo during embryogenesis across a range of animal species. Credit: Bazzini Lab, Stowers Institute
“That maternal contribution is a mystery that many of us want to solve,” says Bazzini. However, attempts to systematically target RNA in zebrafish, the model organism of choice for many developmental biologists, have been unsuccessful. The aptly-named RNA interference method, which has been a mainstay in studies of gene function, does not work in zebrafish, or other fish or frogs. Other methods using synthetic strips of genetic code known as morpholinos or antisense oligonucleotides have sometimes been associated with toxicity and off-target effects.
So when Bazzini and his collaborator and friend Miguel A. Moreno-Mateos, Ph.D., a professor at Pablo de Olavide University, noticed reports that CRISPR technology had been employed to degrade RNA in yeast, plants, and mammalian cells, they were eager to give it a try. Moreno-Mateos was a postdoc in Antonio Giraldez’s lab at Yale University at the same time as Bazzini, and is considered an expert on the optimization of CRISPR-Cas technology in vivo.
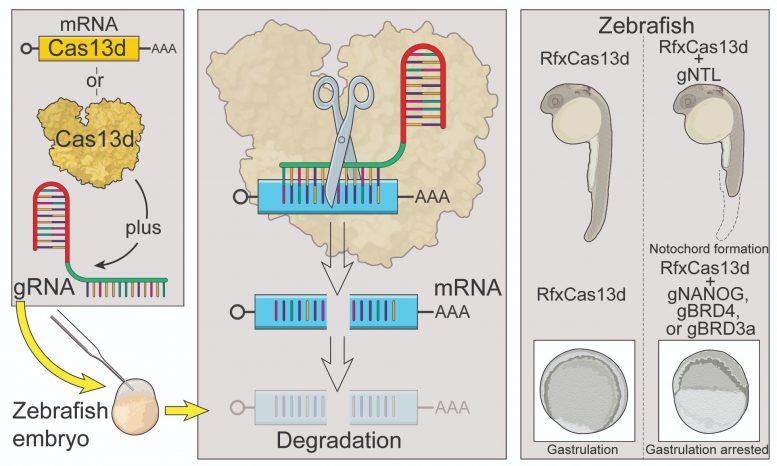
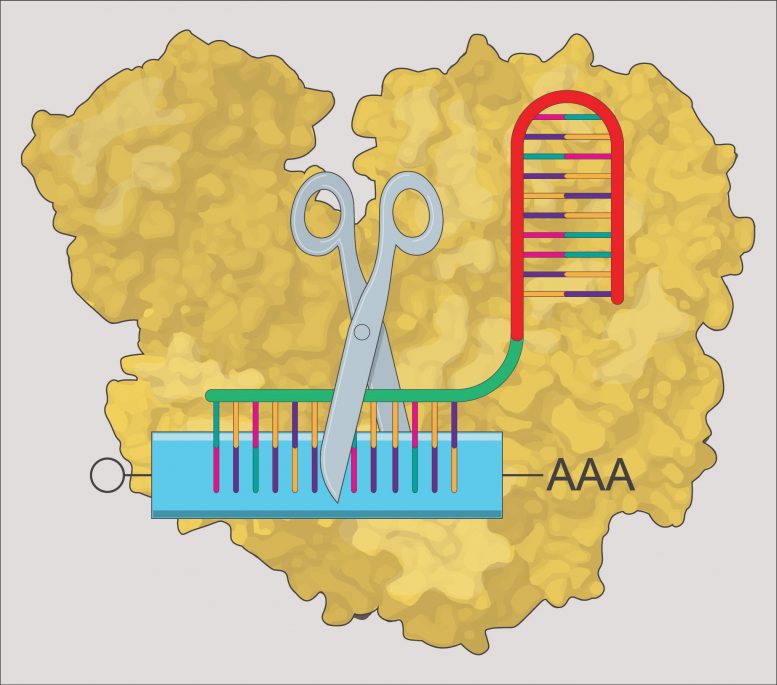
The CRISPR-Cas13 system depends on two ingredients — a short RNA sequence known as a “guide” RNA, and an enzyme called Cas13 (part of the Cas, or CRISPR-associated, family of proteins) that cuts any RNA messages in the cell that could line up and bind to that guide sequence. The researchers tested four different Cas13 proteins that had been successfully used in previous studies. They found that the Cas13 proteins were either inefficient or toxic to the developing zebrafish, except for one protein, called RfxCas13d.
They then examined whether targeting RNA with CRISPR-RfxCas13d in zebrafish embryos could recreate the same defects as altering the organism’s underlying DNA. For example, when they targeted the RNA of the tbxta gene, which is necessary for growing a tail, the zebrafish embryos were tailless.
The researchers went on to show that the CRISPR system could efficiently target a variety of RNAs, both those provided by the mother as well as those produced by the embryo, decreasing RNA levels by an average of 76%. Collaborators within and outside of Stowers helped derive that statistic, and showed that the technique also works in killifish, medaka, and mouse embryos.
“The CRISPR-RfxCas13d system is an efficient, specific, and inexpensive method that can be used in animal embryos in a comprehensive manner,” says Moreno-Mateos, who is also co-leader of the study. “With this tool we will help to understand fundamental questions in biology and biomedicine.”
One of the fundamental questions the researchers hope to pursue is the role that RNA plays in the earliest hours of development. The RNAs left behind by the mother have to be removed at precisely the same time that the genome of the embryo comes online; otherwise, the embryo never develops.
“We think this tool could have a profound effect on our understanding of infertility and developmental problems in general,” says Bazzini.
“The Stowers facilities and collaborative environment have allowed us to test CRISPR technology in other animal model systems,” Bazzini says. “When I joined Stowers about four years ago, I would have never predicted that my lab would be doing experiments in mouse or killifish models. It’s been a fun adventure!”
Reference: “CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos” by Gopal Kushawah, Luis Hernandez-Huertas, Joaquin Abugattas-Nuñez del Prado, Juan R. Martinez-Morales, Michelle L. DeVore, Huzaifa Hassan, Ismael Moreno-Sanchez, Laura Tomas-Gallardo, Alejandro Diaz-Moscoso, Dahiana E. Monges, Javier R. Guelfo, William C. Theune, Emry O. Brannan, Wei Wang, Timothy J. Corbin, Andrea M. Moran, Alejandro Sánchez Alvarado, Edward Málaga-Trillo, Carter M. Takacs, Ariel A. Bazzini and Miguel A. Moreno-Mateos, 7 August 2020, Developmental Cell.
DOI: 10.1016/j.devcel.2020.07.013
Other coauthors from the Stowers Institute include Gopal Kushawah, Ph.D., Michelle DeVore, Huzaifa Hassan, Wei Wang, PhD, Timothy J. Corbin, Andrea M. Moran, and Alejandro Sánchez Alvarado, Ph.D.
This research was funded by the Stowers Institute for Medical Research, Pablo de Olavide University, Consejo Superior de Investigaciones Cientificas, and Junta de Andalucia. Additional support included the Ramon y Cajal program (RyC-2017-23041) and grants (BFU2017-86339-P, PGC2018-097260-B-I00, and MDM-2016-0687) from the Spanish Ministerio de Ciencia, Innovación y Universidades; the Springboard program from Centro Andaluz de Biología del Desarrollo; Genome Engineer Innovation 2019 Grant from Synthego; the Pew Innovation Fund; Innovate Peru (grant 168-PNICP-PIAP-2015); and FONDECYT (travel grant 043-2019).






Be the first to comment on "Scientists Use CRISPR to Target Gene Messages Involved in Early Development"