
A groundbreaking study employing full genome-scale data has revolutionized our understanding of bird evolution, revealing new insights into the phylogenetic relationships among bird species. By analyzing a comprehensive dataset of 363 bird species, the research has redefined the avian family tree, identifying four major clades within Neoaves and shedding light on the complex evolutionary history of birds. Credit: SciTechDaily.com
The most extensive genomic study to date has unveiled how the birds spread all over the world after mass extinction.
Birds represent the sole surviving lineage of dinosaurs in the present day. Roughly 66 million years ago, at the transition from the Cretaceous to the Paleogene period (the K–Pg boundary), a catastrophic extinction event led to the demise of all non-avian dinosaurs, providing an opportunity for birds to diversify rapidly and occupy a wide range of ecological niches.
Neoaves, a diverse group comprising approximately 95% of all bird species today, emerged from this radiation. From the towering condors of the Andes to the diminutive hummingbirds flitting through tropical forests, Neoaves encompass a stunning diversity of forms and functions.
Despite considerable efforts to reconstruct avian evolutionary history and the impacts of the K-Pg event using morphological data and molecular data, the precise branching order and relationships among the neoavian lineages remained contentious.
“Previous studies on small datasets from different genomic regions often produced conflicting results with respect to the topology of bird tree,” said Guojie Zhang, senior author of the paper, a chair professor on evolutionary biology in Zhejiang University, and one of the initiators of the B10K project. “In this study, for the first time, we employed full genome-scale data to construct the tree for bird species from almost all representative families.”
Advancements in Genomic Data
This full genomic dataset was produced by the B10K consortium in its second phase which includes 363 bird species covering all major bird lineages.
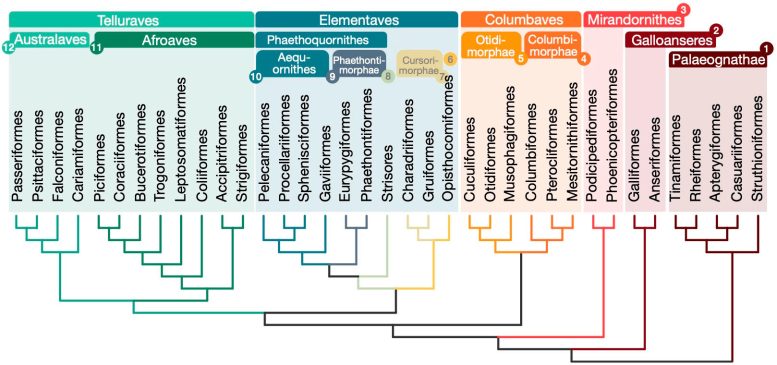
The new family tree is breaking new ground in the long journey to unravel the mysteries of bird evolution. According to this updated bird family tree, a group containing flamingos and grebes (called Mirandornithes) were among the first neoavian lineages to evolve.
The new tree is challenging the organization of Neoaves by classifying this big group into four major clades: Mirandornithes, Columbaves, Elementaves, and Telluraves. Elementaves is a newly proposed grouping comprising ca. 14% of all species of modern birds including disparate groups such as the enigmatic hoatzin, shorebirds, hummingbirds, and tropicbirds.
Elementaves is named to reflect the group’s remarkable diversity in ecological niches, representing the major elements of Earth, air, and water. This new family tree resolves some long-standing debates over the relationships among avian species and lays a solid foundation for studying avian evolution and trait development.
The Impact of Comprehensive Genomic Analysis
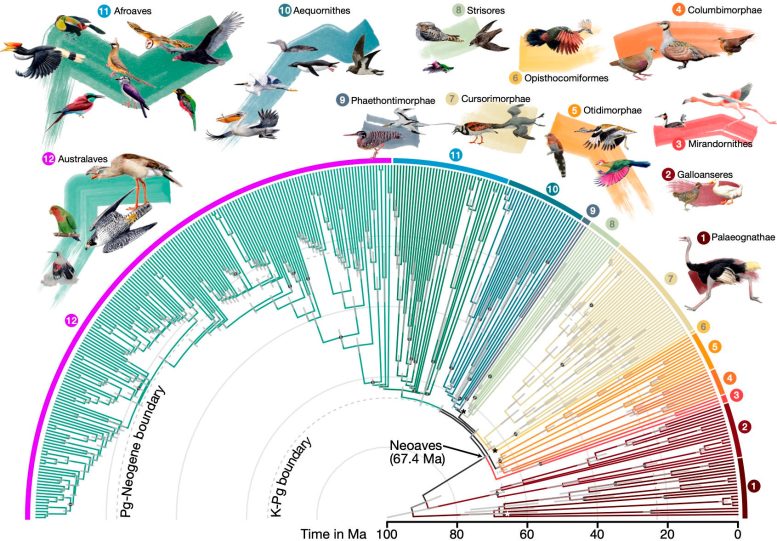
By employing full genome data across 363 bird species, this is the largest-ever dataset used for phylogenetic analyses of birds. The team built a new pipeline to extract over 150,000 regions spread out across the genome.
“We characterized phylogenetic relationships across the entire genome and identified patterns associated with the genomic context and sequence characteristics,” said Josefin Stiller, the leading author of this study and an Assistant Professor in evolutionary biology at the University of Copenhagen, “We found that various parts of the genome, for example, individual chromosomes or protein-coding genes, often support vastly different trees. This likely explains why studies that only analyzed certain genomic parts were in conflict.”
Putting together high-quality and large amounts of data was key to producing a robust phylogenetic tree. The team discovered that for most branches, a consensus on their relationships can be reached when a sufficient amount of data was used. But the phylogenetic positions for some bird groups like owls and hawks remain puzzling even with a full-genome scale of data.
“More data does not necessarily produce a better solution,” Zhang said. Siavash Mirarab, co-senior author of the study, a professor of electrical and computer engineering at the University of California, San Diego, added that, “The reason for this may be some complex evolutionary history like ancient cross-mating between two lineages, incomplete lineage sorting, long-branch attraction, and biased DNA sequence content, all of which can interfere with the reconstruction of phylogenetic trees.”
The study reports new insights on which of these factors impact which branches of the tree, providing a more comprehensive and genuine picture of the origin of these avian groups. The study also proposes a more accurate time scale for the diversification of modern birds, suggesting that a rapid radiation occurred at or near the mass extinction at the Cretaceous–Paleogene (K–Pg) boundary and to a lesser degree shortly after the Paleogene–Neogene boundary.
The researchers found that these radiations coincided with remarkable genetic and morphological changes among birds, including greater mutation rates, smaller body sizes, and larger brains, and larger effective population sizes. “This illustrates the power of comparative genomics: by comparing genomes of living species, we can uncover traces of events that happened 66 million years ago,” Stiller said.
“Our work has changed many traditional views on the evolutionary history of birds. This new family tree will serve as a solid backbone for mapping the evolutionary history of all bird species with important implications for ornithological research and biodiversity studies,” Zhang concluded.
Reference: “Complexity of avian evolution revealed by family-level genomes” by Josefin Stiller, Shaohong Feng, Al-Aabid Chowdhury, Iker Rivas-González, David A. Duchêne, Qi Fang, Yuan Deng, Alexey Kozlov, Alexandros Stamatakis, Santiago Claramunt, Jacqueline M. T. Nguyen, Simon Y. W. Ho, Brant C. Faircloth, Julia Haag, Peter Houde, Joel Cracraft, Metin Balaban, Uyen Mai, Guangji Chen, Rongsheng Gao, Chengran Zhou, Yulong Xie, Zijian Huang, Zhen Cao, Zhi Yan, Huw A. Ogilvie, Luay Nakhleh, Bent Lindow, Benoit Morel, Jon Fjeldså, Peter A. Hosner, Rute R. da Fonseca, Bent Petersen, Joseph A. Tobias, Tamás Székely, Jonathan David Kennedy, Andrew Hart Reeve, Andras Liker, Martin Stervander, Agostinho Antunes, Dieter Thomas Tietze, Mads Bertelsen, Fumin Lei, Carsten Rahbek, Gary R. Graves, Mikkel H. Schierup, Tandy Warnow, Edward L. Braun, M. Thomas P. Gilbert, Erich D. Jarvis, Siavash Mirarab and Guojie Zhang, 1 April 2024, Nature.
DOI: 10.1038/s41586-024-07323-1






Be the first to comment on "From Dinosaurs to Hummingbirds: New Family Tree Revises Our Understanding of Bird Evolution"