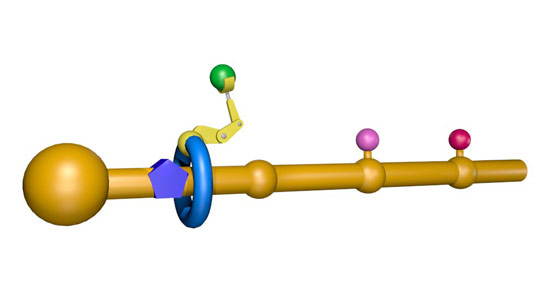
With molecules for moving parts, this nanorobot links together amino acids (colored balls) by attaching them to a moving ring (blue). Credit: Miriam Wilson
Scientists have invented a nanomachine that mimics the function of the ribosome, which is the molecular machine that translates the genetic code into the body’s proteins.
Dave Leigh, a synthetic chemist at the University of Manchester, UK, and his team published their findings in the journal Science¹. The machine is much simpler than the ribosome, and at about one-tenth of the size and very slow. It destroys the code it reads and only produces short chunks of peptides, but some of these tactics could be used to make useful chemicals.
The machine built by Leigh and his team relies on a rotaxane, a large molecular ring threaded into another molecule that acts like an axle, which is lined with three amino acids. A chain of three amino acids hangs from the outer edge of the ring. One of these amino acids is cysteine, which contains a crucial sulfur-containing thiol group.
When the system is heated up, the thiol group plucks an amino acid from the axle and transfers it to the end of the chain. The ring moves along the axle and repeats this process again. Unthreading the ring and breaking off the newly-formed chain delivers a peptide made from all six amino acids.
Professor Leigh’s molecular machine is based on the ribosome. It features a functionalized nanometre-sized ring that moves along a molecular track, picking up building blocks located on the path and connecting them together in a specific order to synthesize the desired new molecule. First the ring is threaded onto a molecular strand using copper ions to direct the assembly process. Then a “reactive arm” is attached to the rest of the machine and it starts to operate. The ring moves up and down the strand until its path is blocked by a bulky group. The reactive arm then detaches the obstruction from the track and passes it to another site on the machine, regenerating the active site on the arm. The ring is then free to move further along the strand until its path is obstructed by the next building block. This, in turn, is removed and passed to the elongation site on the ring, thus building up a new molecular structure on the ring. Once all the building blocks are removed from the track, the ring de-threads and the synthesis is over. Credit: Miriam Wilson
Leigh’s technique of sulfur-assisted amino acid transfer is found elsewhere in biology. Some bacteria use sulfur-assisted amino acid transfers to synthesize proteins. It’s also commonly used in protein synthesis in the lab.
When 1018 of Leigh’s molecular machines are run at once, tens of milligrams of the peptide are produced, but it is still a slow process, taking roughly 12 hours to attach each amino acid in the sequence. The ribosome, on the other hand, attaches 15 to 20 amino acids per second.
In its current form, the nanomachine requires the axle to be preloaded with amino acids in the correct sequence and can assemble just one peptide. In contrast, the ribosome grabs amino acids out of the liquid medium and assembles them in the right order according to a template. The team hopes to develop a molecular machine that does something similar in the future.
The goal is to move chemistry beyond its usual habit of making molecules using stepwise processes. These machines could eventually allow chemists to build materials with a specific sequence of molecules.
Reference: “Molecular robot mimics life’s protein-builder” by Mark Peplow, 10 January 2013, Nature.
DOI: 10.1038/nature.2013.12190








Be the first to comment on "Molecular Robot Mimics the Ribosome"